Mechanism of transcriptional bursting in bacteria

Mechanism of TranscriptionalBursting in Bacteria
Shasha Chongyi Hao Ge,and X. Sunney 1Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA 02138, USA2Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138, USA3Biodynamic Optical Imaging Center (BIOPIC), Peking University, Beijing 100871, China4Beijing International Center for Mathematical Research (BICMR), Peking University, Beijing 100871, China5Co-first author*Correspondence:
ported a high-throughput, single-molecule fluorescence in situhybridization (FISH) assay to measure the cellular copy number
Transcription of highly expressed genes has been
distribution of a particular mRNA for a large population of
shown to occur in stochastic bursts. But the origin
isogenic E. coli cells ). When mRNAs are
of such ubiquitous phenomenon has not been
generated with a constant flux, one expects a Poisson distribu-
understood. Here, we present the mechanism in
tion of mRNAs across the population. Bursting transcription
bacteria. We developed a high-throughput, in vitro,
would lead to non-Poissonian distributions. For all the highlyexpressed
single-molecule assay to follow transcription on
E. coli genes, we found that the distributions are not
Poissonian, with the Fano factor (variance divided by the mean
individual DNA templates in real time. We showed
of a given distribution) larger than one. This indicates the ubiquity
that positive supercoiling buildup on a DNA seg-
of transcriptional bursting in bacteria.
ment by transcription slows down transcription
However, the origin of bacterial transcriptional bursting is still
elongation and eventually stops transcription initia-
unknown. Its stochasticity implies it is a single-molecule
tion. Transcription can be resumed upon gyrase
behavior: there is only one copy of the gene in the cell. Its univer-
binding to the DNA segment. Furthermore, using
sality implies that it cannot be attributed to a specific gene or
single-cell mRNA counting fluorescence in situ hy-
protein factor. Rather, it must originate from a fundamental and
bridization (FISH), we found that duty cycles of
general mechanism pertinent to the chromosomal DNA structure
transcriptional bursting depend on the intracellular
and its influence on transcription regulation.
gyrase concentration. Together, these findings
It has been shown that E. coli chromosomal DNA is segre-
gated to �400 topologically constrained loops with an average
prove that transcriptional bursting of highly ex-
size of 10,000 base pairs (
pressed genes in bacteria is primarily caused by
). Recent work discussed that E. coli nucleoid-asso-
reversible gyrase dissociation from and rebinding
ciated proteins such as H-NS and Fis can cause formation of
to a DNA segment, changing the supercoiling level
DNA loops based on both chromosome conformation capture
of the segment.
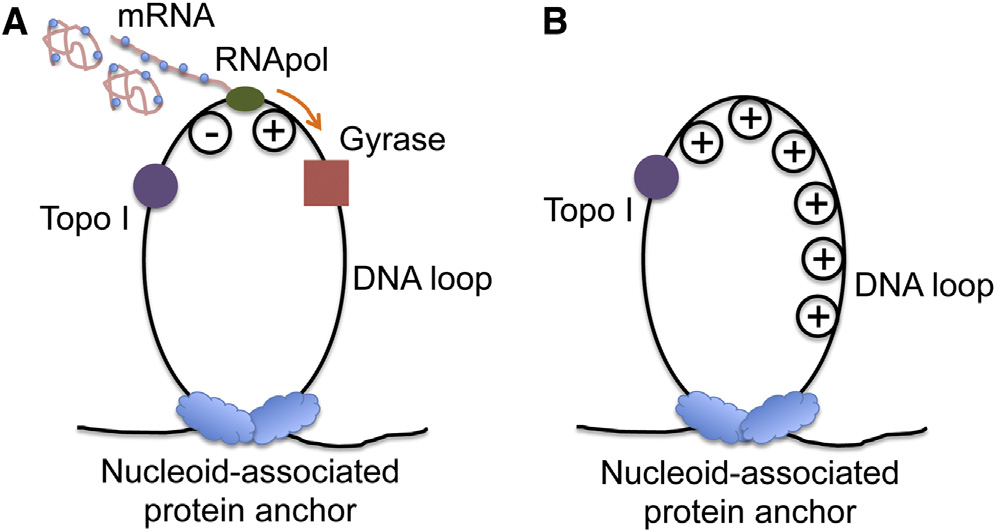
and superresolution optical imaging experiments (). Such chromosome structure provides us a clue to explain
the transcriptional bursting phenomenon (In such aDNA loop, transcription generates positive supercoiling ahead
Essential for all cell functions, transcription, the synthesis of
of the RNApol and negative supercoiling behind the RNApol
mRNAs from DNA carried out by RNA polymerase (RNApol), is
the first step in gene expression. Many recent experiments
There exist two major
have shown the general phenomenon that transcription of
topoisomerases in E. coli cells, gyrase and topoisomerase I
highly expressed genes occurs in stochastic bursts in bacteria
(Topo I), which release positive and negative supercoiling,
respectively It is known that negative supercoiling
and eukaryotic cells (A major
formed during transcription elongation is rapidly removed by
source of gene expression noise, transcriptional bursting results
Topo I (This is necessary because accu-
in cellular diversity of an isogenic population, possibly enhancing
mulation of negative supercoiling could lead to the formation
survival of the population in the face of environmental uncertainty
of detrimental R loops, an RNA-DNA hybrid (The
activity of gyrase, on the other hand, is not as sufficient to
Golding and coworkers directly observed tran-
keep up with transcription (), leading to posi-
scriptional bursting in real time by using MS2 loops to monitor
tive supercoiling accumulation on the DNA loops containing
mRNA production in E. coli (Our group re-
highly transcribed operons
314 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
Therefore, we were able to track transcription elongationin real time as the nascent mRNA being produced on a sur-face-tethered DNA template. Transcription activities on up tohundreds of templates in one field of view can be monitoredsimultaneously.
As a control, we examined the effect of SYTO RNASelect on
the activities of enzymes involved in our system, including T7RNApol, E. coli RNApol, E. coli gyrase, and E. coli Topo I.
None of them were found to be affected by the stain (FiguresS1A–S1H available online). With sufficiently low laser powerand the presence of a fresh oxygen scavenger system, photo-bleaching of the dye and photocleavage of nucleic acids werenegligible (Figure S2).
Figure 1. Transcription on Topologically Isolated Chromosomal
In our single-molecule assay, DNA templates containing a pro-
moter were tethered on the passivated surface of the flow cell
(A) Gyrase releases positive supercoiling generated by transcription on a DNA
through biotin-streptavidin linkage. After we flowed RNApol
loop, and RNApol keeps transcribing the gene.
and nucleoside triphosphates (NTPs) into the flow cell, the fluo-
(B) In the absence of gyrase, active transcription on a DNA loop leads to
rescence intensity of many spots in the field of view linearly
positive supercoiling accumulation, which inhibits further transcription on theparticular DNA loop.
ramped up due to transcription elongation, followed by abruptdisappearance upon transcription termination (‘‘Blinking'' of fluorescence occurred when multiple transcriptswere produced. As a control, no fluorescence intensity increase
It has been found that there are �500 gyrase molecules per
was observed under any of the following conditions: (1) no
E. coli cell
RNApol in the solution, (2) no NTPs in the solution, and (3) no pro-
which happens to be roughly the number of topo-
moter in the DNA template. Full-length transcripts (>12 kb) were
logically constrained DNA loops per chromosome. On average,
generated as confirmed by RNA gel electrophoresis (Figures
there is one gyrase molecule per DNA loop. When a gyrase
molecule reacts on the DNA loop, positive supercoiling is
By recording fluorescent movies, we were able to measure
released, and RNApol can keep transcribing the gene (‘‘on''
intensity versus time for a field of view containing hundreds of
state; When the gyrase dissociates from the loop,
individual DNA templates, from which we could monitor how
positive supercoiling is built up by transcription, possibly slow-
individual transcripts were generated (This in vitro,
ing down transcription elongation and stopping transcription
single-molecule assay allows us to investigate the effects of
initiation (‘‘off'' state;
supercoiling on transcription initiation and elongation in a clean
In this study, through a series of in vitro single-molecule and
and controlled system.
live-cell experiments, we prove that transcriptional bursting ofhighly expressed genes in bacteria is primarily caused by gyrase
Positive Supercoiling Buildup by Transcription Slows
dissociation from and reversible binding to a DNA segment or a
Down Transcription Elongation
chromosomal loop, which changes the supercoiling level of the
We examined the effect of positive supercoiling buildup on
DNA segment.
transcription elongation in vitro. We designed 12-kb-longlinear DNA templates with T7 or E. coli promoter on the 50 end
and single or multiple biotinylated nucleotides on the 30 end(
An In Vitro, Single-Molecule Assay Allows Real-Time
When the DNA duplex is tethered to the surface with a single
Monitoring of Transcription on Individual DNA
biotin-streptavidin linkage, we found the average T7 transcrip-
tion elongation rate is 53.2 ± 3.4 nt/s (0.3 mM each NTP;
We developed an in vitro, single-molecule assay to monitor re-
23�C), which is consistent with previously reported rates
petitive stochastic transcription events in real time on individual
). This result further proved that transcription was not
DNA templates with controlled supercoiling levels (A).
affected by the SYTO RNASelect dye. In this case, supercoiling
We used a nucleic acid stain, SYTO RNASelect (Life Technolo-
cannot accumulate because DNA can rotate around its single
gies), which is nonfluorescent at 530 nm but becomes fluores-
linkage to the surface.
cent upon binding to RNA (B). It has been used to detect
On the other hand, DNA with multiple biotinylated nucleotides
RNA in the presence of DNA (). An argon
cannot rotate around its multiple linkages to the surface. Positive
laser line at 488 nm was used to excite SYTO RNASelect in
supercoiling would accumulate downstream of the elongation
a total internal reflection fluorescence (TIRF) microscope. We
complex when spiral of the bulky complex around the DNA is
collected fluorescence at 530 nm and recorded time-lapse
hindered by the frictional drag on the complex. Interestingly,
movies with a charge-coupled device (CCD) camera. In the
we found T7 transcription elongation was slowed down by
presence of the dye, a single nascent mRNA becomes visible,
38% as positive supercoiling accumulated on the multiple-biotin
and its fluorescence intensity increases with the mRNA length.
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 315
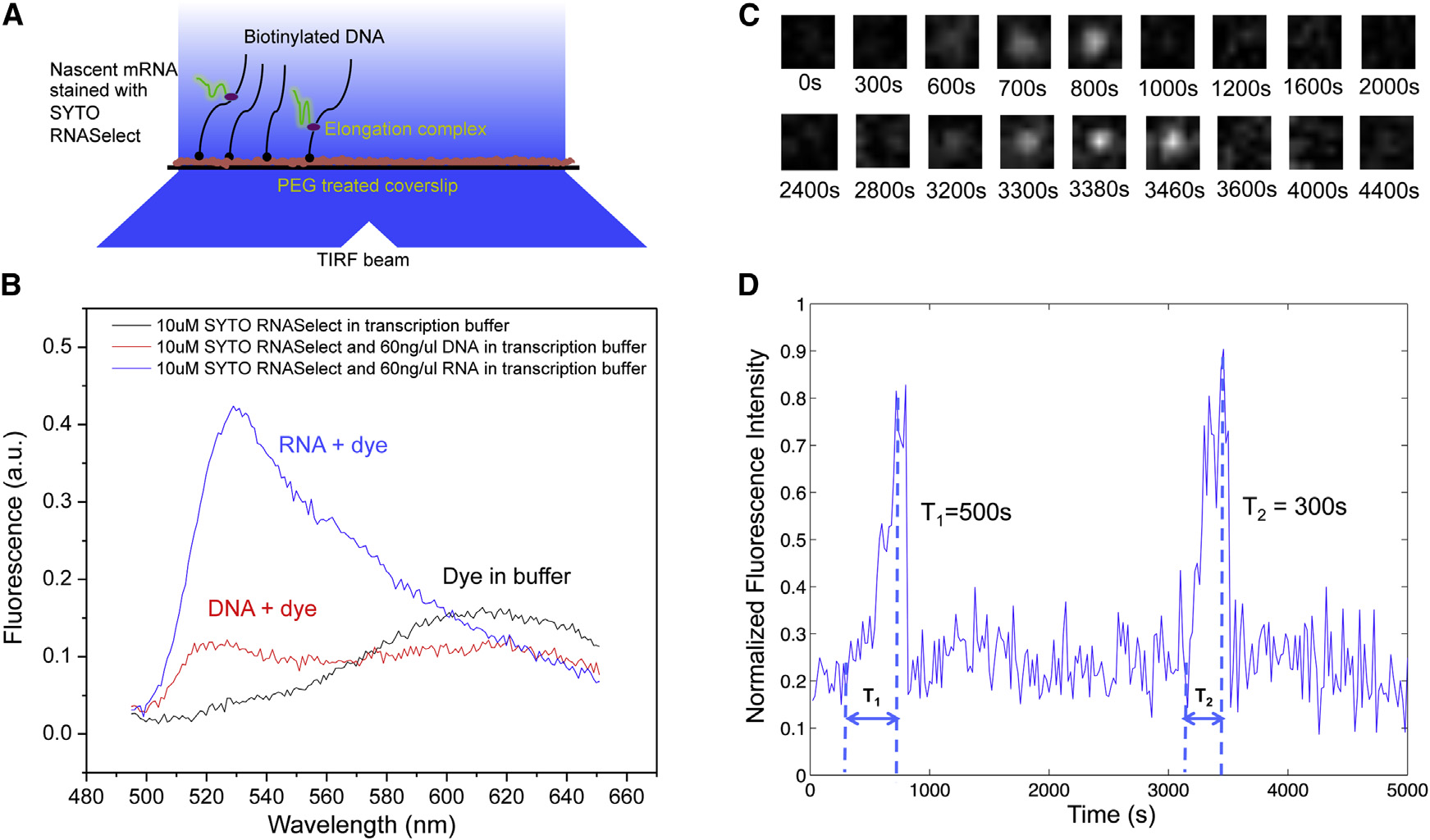
Figure 2. In Vitro, Single-Molecule Assay to Monitor Real-Time Transcription on Individual DNA Templates Using SYTO RNASelect Stain(A) Schematic representation of the experimental arrangement (not drawn to scale). In the presence of 250 nM SYTO RNASelect, nascent RNAs are fluorescentunder TIRF excitation at 488 nm. With an excitation power density of 0.22 W/cm2 and an image acquisition time of 5 s, a transcript of 2,300 nucleotides yields asignal-to-noise ratio of 1.
(B) Fluorescence emission spectra of SYTO RNASelect solution under 488 nm excitation. The dye selectively stains RNA and emits fluorescence with a peak at530 nm. In the absence of nucleic acids, the dye is not fluorescent at 530 nm. a.u., arbitrary units.
(C) Time-lapse images of 1.1 3 1.1 mm sub-field of view to monitor T7 transcription on one 12-kb-long template.
(D) Intensity-versus-time trajectory of the DNA template shown in (C). Full transcripts are produced repetitively on the template, with transcription elongation timeT1 = 500 s and T2 = 300 s, respectively.
See also Figures S1 and S2.
There is a concern that supercoiling might arise from immo-
Dissociation Constant of Gyrase-DNA Complex Is
bilization of the elongation complex to the surface. Special
Determined from Gyrase Concentration Dependence of
care was taken to minimize interactions of the elongation com-
Transcription Elongation Rates
plex with the surface in our experiment. Besides, we note our
Gyrase-DNA binding can be described by two steps
result is consistent with the earlier in vitro report that the fric-
). First, DNA and gyrase form a complex with limited pro-
tional drag on a sizable nascent transcript is enough to lead
tein-DNA-binding interface, which is prone to rapid dissociation.
to DNA supercoiling (even in aqueous solu-
Second, a chiral DNA wrap is formed around gyrase, which in the
tion and free of surface perturbation. Moreover, the measured
presence of ATP generates negative DNA supercoils. Here, we
elongation rate with the single linkage did not seem to be
discuss the binding stability and kinetics of the DNA wrapping
perturbed by the surface interaction with the elongation com-
state, which are relevant to transcription dynamics.
plex, if any.
By titrating the elongation rate on the multiple-biotin template
Interestingly, the elongation rate on the multiple-biotin tem-
with gyrase (we determined the gyrase-DNA dissoci-
plate was recovered when gyrase was added into the system.
ation constant Kd from the gyrase concentration at which the in-
shows the elongation rate as a function of gyrase con-
crease of the T7 transcription elongation rate reaches half of its
centration, reaching the value of the single-biotin template at a
saturation value, that is Kd z 100 nM (
saturating gyrase concentration. As a control, we found that
This Kd is larger than previously reported
gyrase did not affect the elongation rate on the single-biotin tem-
plate indicating that gyrase play no other role than
), where specific gyrase-binding sequences were used
releasing positive supercoiling.
Similarly, with E. coli RNApol, we found positive supercoiling
gyrase-binding sites comparable to these sequences are
accumulation on the multiple-biotin template also slowed
sparsely distributed on the E. coli chromosome with a frequency
down transcription elongation by 47% (which is
of only one per 100 kb ). The nuoB-N
consistent with a recent report based on mechanical manipula-
DNA sequence (�12 kb) we used in our in vitro assay better rep-
resents a chromosomal DNA loop (�10 kb) that binds to gyrase
316 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
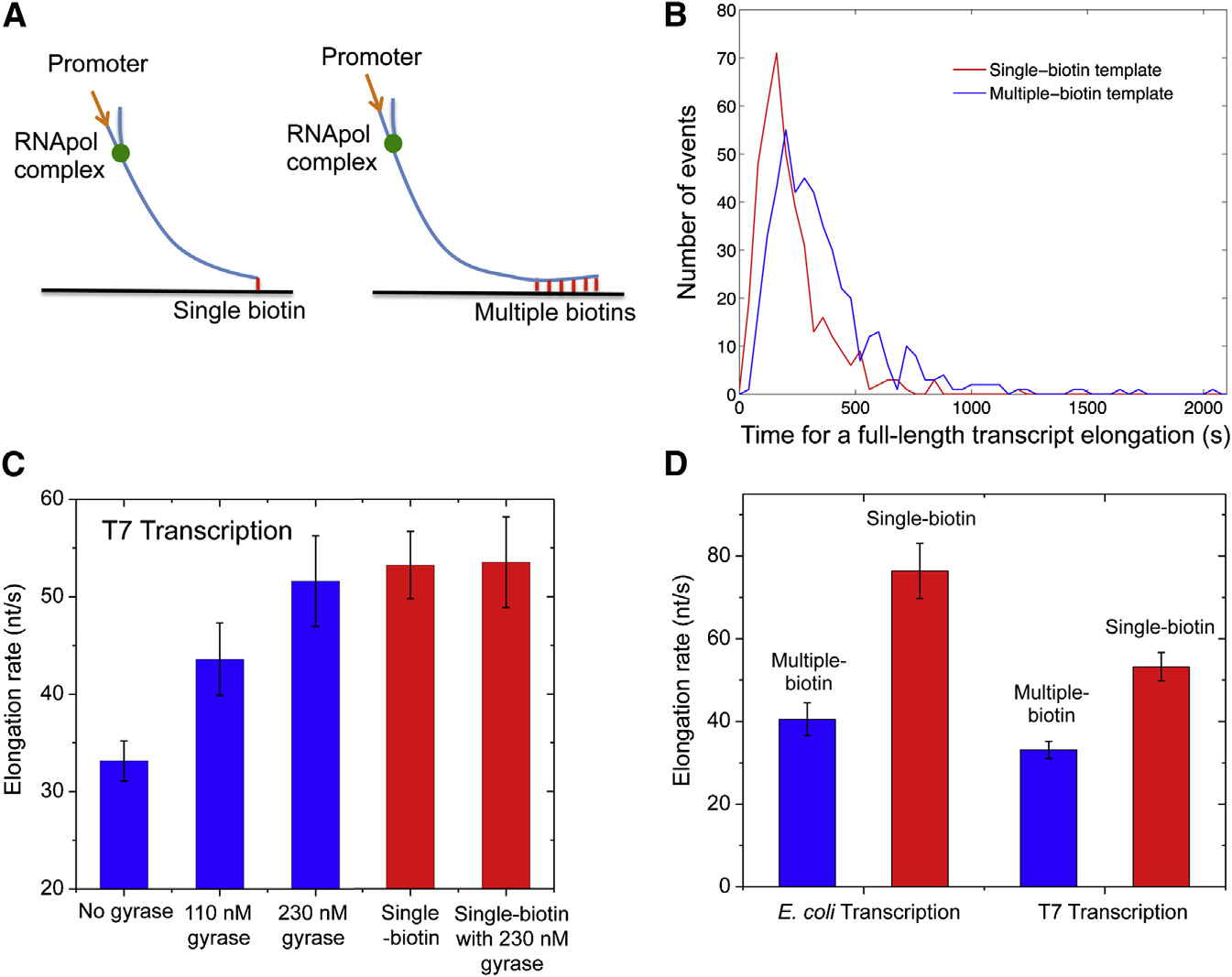
Transcription Elongation Rate(A) In vitro transcription template design contain-ing a T7 or T7A1 promoter and a 12 kb transcribingsequence. The template is anchored to the flowcell surface via either a single or multiple biotin-streptavidin linkages.
(B) Histogram of T7 transcription elongation timeon the templates anchored with single (red curve)or
curve). The average elongation time for the multi-ple-biotin template is 60% longer.
(C) Titration of T7 transcription elongation rate(23�C) on the multiple-biotin template (the threebars on the left) with gyrase concentration. Theelongation rate increases with the gyrase con-centration till it gets as high as that on the single-biotin template (the fourth bar). The elongation rateon the single-biotin template does not change inthe presence of a saturating concentration ofgyrase (the bar on the right).
(D) E. coli transcription elongation rate (37�C) andT7 transcription elongation rate (23�C) on themultiple-biotin template are slower than on thesingle-biotin template. In (C) and (D), the elonga-tion rates are averaged from over 300 transcriptsunder each condition. The error bars are boot-strapped confidence intervals See also Figures S1 and S2.
at multiple weak binding sites
nation A). We found the initiation rate was indeed con-
stant over time because of repetitive annihilation of supercoiling(
Positive Supercoiling Buildup by Transcription
We then examined the second steady-state condition, in
Essentially Stops Transcription Initiation
which T7 transcription occurs on the circular templates in
Next, we examined the effect of positive supercoiling on tran-
the presence of both Topo I and gyrase C). Because
scription initiation. In order to mimic topologically isolated DNA
both positive and negative supercoiling on the DNA template
loops in the bacterial chromosome, we designed a circular tem-
is continuously removed, the initiation rate remained constant
plate (A) and tethered it to the surface with multiple
over time under this condition (D), which is the
biotin-streptavidin linkages. The circular template consists of a
same as that in the first steady-state condition (Figures S4C
T7 or E. coli promoter, a 12-kb-long transcribing sequence,
and a T7 or E. coli terminator. Due to the low circularization effi-
We now examine how positive supercoiling buildup would
ciency, a significant fraction of the purified DNAs remained to be
hinder transcription initiation. After introduction of Topo I, nega-
linear, which are also tethered on the flow cell surface and tran-
tive supercoiling is rapidly removed, and positive supercoiling is
scribed. We picked the circular templates for analysis by staining
expected to accumulate on the circular template as multiple
the DNA molecules with SYTOX Orange and imaging them under
transcripts are made (E). Indeed, we observed the
flow after recording transcription movies (Figure S3).
initiation rate decreased over time F). Interestingly,
For a single template under steady-state condition, transcrip-
the final intensity has dropped to under 20% of its initial value,
tion initiation rate is the number of initiation events over a fixed
indicating that transcription initiation was essentially stopped
period of time (frequency of ‘‘spikes'' in the intensity trajectory
by the buildup of positive supercoiling. This final state corre-
from a template; Figure S4A). According to the ergodic principle,
sponds to the gene ‘‘off'' state. We found that it takes approx-
the initiation rate is the sum of initiated events from a population
imately nine rounds of T7 transcription to build up sufficient
of templates at a specific time point. We measured the total
positive supercoiling that inhibits transcription initiation on a
intensity of the circular templates, which is proportional to the
single template in vitro (Figure S4B and
initiation rate.
We examined the first steady-state condition, in which T7
Similar to T7 transcription, we found transcription initiation
transcription occurs on the circular templates in the absence of
rate of E. coli RNApol dropped to �25% after approximately
Topo I and gyrase. A bulky elongation complex generates
five transcripts were produced from a circular template of the
positive supercoiling ahead of it and negative supercoiling
same length (12 kb) in the presence of Topo I E, 4G,
behind it, which annihilate each other when the elongation
and S5; ). We note that
complex dissociates from the template upon transcription termi-
fewer than five rounds of transcription might be sufficient to
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 317
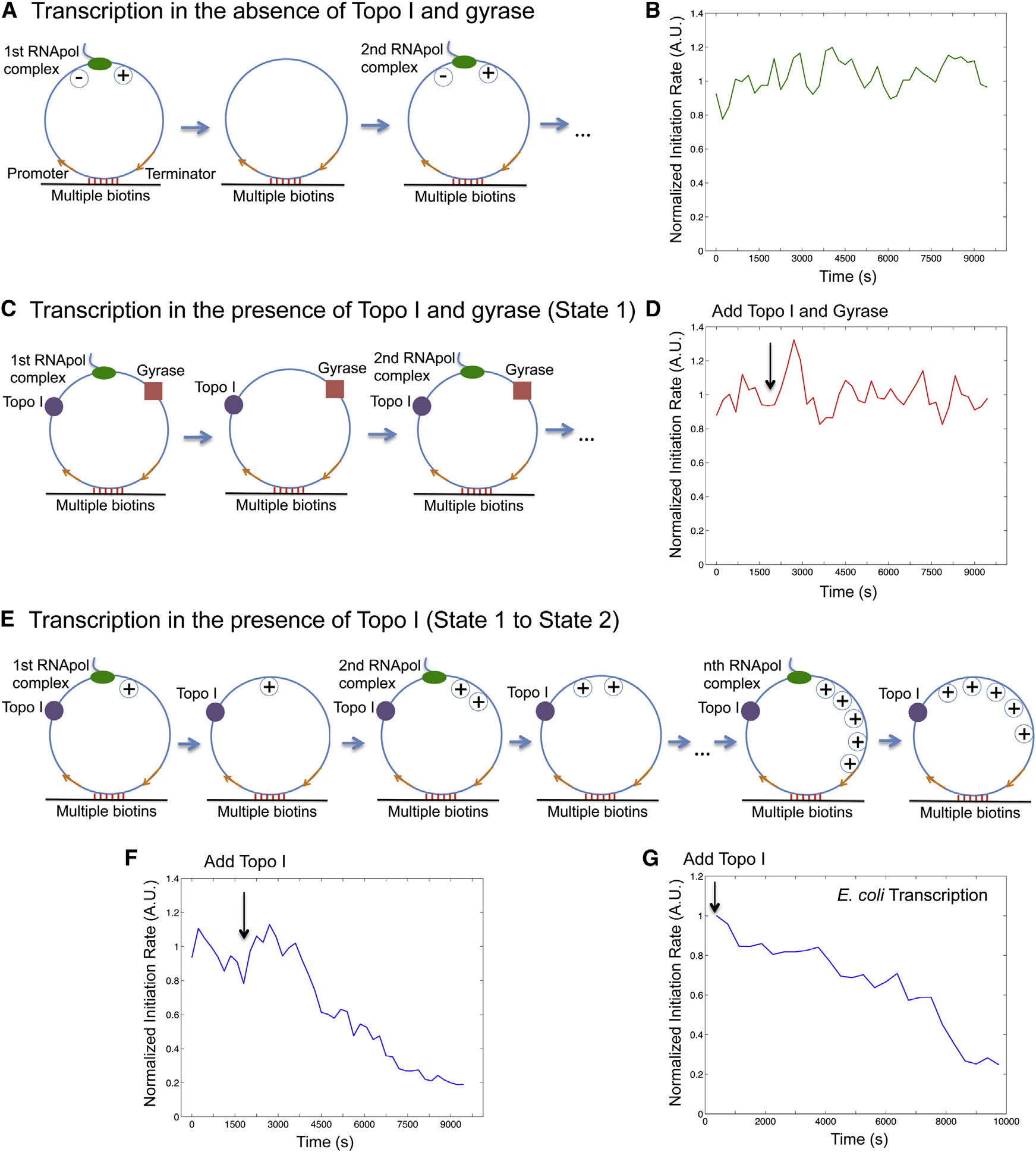
Figure 4. Supercoiling Dependence of Transcription Initiation Rate(A) Schematic of transcription on a circular template in the absence of topoisomerases. Positive and negative supercoiling annihilate each other after RNApolcompletes transcription and dissociates from the template.
(B) Time dependence of T7 transcription initiation rate under the condition of (A).
(C) Schematic of transcription on the circular template in the presence of 41 nM Topo I and 0.1 mM gyrase (same as state 1 in (D) Time dependence of T7 transcription initiation rate under the condition of (C). The arrow shows the time when the topoisomerases were added into the system.
(E) Schematic of transcription on the circular template in the presence of 41 nM Topo I and absence of gyrase. Positive supercoiling is built up as transcripts areproduced.
(F) Time dependence of T7 transcription initiation rate under the condition of (E). (B), (D), and (F) are the total intensity versus time from 160 circular templatesunder respective conditions normalized to the same fluorescence intensity.
(G) Time dependence of E. coli transcription initiation rate in the presence of 62 nM Topo I and absence of gyrase. This is the intensity averaged from 106 circulartemplates at each time point normalized to that from 209 linear templates ).
See also Figures S1, S2, S3, S4, and S5.
318 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
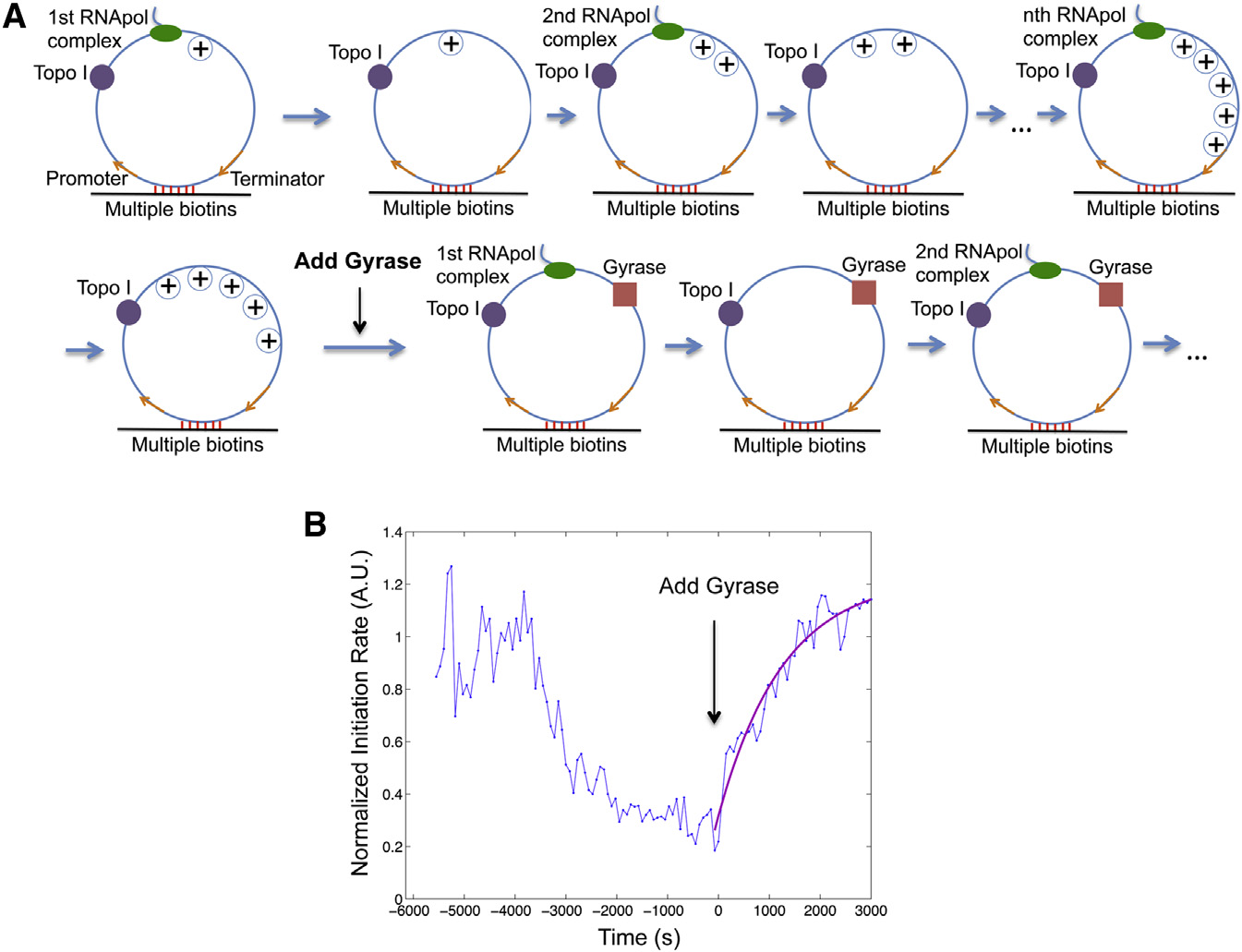
Figure 5. Transition from Gene Off to OnState(A) Schematic of transcription on the circulartemplate first in the presence of 41 nM Topo I only(same as and then both 41 nM Topo Iand 0.1 mM gyrase (same as C).
(B) Time dependence of T7 transcription initiationrate (blue) under the condition of (A). This is theintensity averaged from 160 circular templates ateach time point normalized to that from 120 lineartemplates. Gyrase was added into the system atT = 0, when transcription initiation was essentiallystopped by positive supercoiling accumulation.
The trajectory after T = 0 is fitted with a singleexponential function (magenta).
See also Figures S1, S2, and S3.
concentration used in our in vitro assay0.1 mM. Such consistency suggests thatgyrase binding to the DNA template isthe rate-limiting step to restart transcrip-tion in vitro.
The Kd and kon of gyrase-DNA binding
determined in vitro allow us to estimatethe time it takes gyrase to dissociatefrom and rebind to a specific chromo-
generate the same level of supercoiling in a live cell, where the
somal DNA loop in an E. coli cell. Because there are compara-
environment is more viscous and the elongation complex is
ble numbers of gyrase molecules and chromosomal DNA loops
bulkier due to transcription-translation coupling (
per E. coli cell, many gyrase molecules are trapped on DNA
loops. According to Kd, the intracellular concentration of un-
With regards to why supercoiling stops transcription initia-
bound gyrase [G] is �0.3 mM
tion, earlier magnetic tweezer experiments have shown that
). Because kon of gyrase-DNA binding is �104 M�1 s�1
DNA positive supercoiling leads to significantly slower and
as determined in vitro, the in vivo pseudo-first-order rate con-
less stable formation of E. coli RNApol-promoter open com-
stant for gyrase-DNA binding kon,[G] is �3 3 10�3 s�1. There-
plex (Therefore, we conclude that the ob-
fore, the average gyrase rebinding time is 1/(kon,[G]) z 6 min.
served inhibition of transcription initiation arises from hindered
Because the dissociation rate constant of gyrase-DNA complex
formation of RNApol-promoter open complex due to positive
is koff = Kd,kon z 10�3 s�1, the average gyrase dissociation
time is 1/koff z 17 min. The gyrase rebinding and dissociationtime is in the same order of magnitude with the off and on pe-
Gyrase Binding to Positively Supercoiled DNA Restarts
riods of transcriptional bursting observed in live E. coli cells
We now prove that gyrase binding on the positively super-
In summary, the in vitro experiments demonstrated that
coiled DNA restarts transcription. We started with T7 tran-
DNA positive supercoiling generated by transcription slows
scription on the circular templates in the presence of Topo I,
down both transcription initiation and elongation and eventually
generating the gene off state Upon addition of
stops initiation. Inhibited transcription initiation and elongation
gyrase into the system, transcription initiation rate started to
can be recovered upon gyrase binding to DNA. Therefore,
increase and reached a plateau at the initial value of the
accumulation and removal of positive supercoiling of a chro-
relaxed templates B), indicating that transcription
mosomal DNA loop containing a highly expressed gene can
initiation was fully recovered when positive supercoiling was
switch the gene off and on. Next, we performed live-cell experi-
released by gyrase.
ments to further support this mechanism.
The initiation rate versus time after the introduction of gyrase
can be fitted well with a single exponential rise
Live-Cell Experiments Confirm that Positive
suggesting a single step is rate limiting for the transition. The
Supercoiling Buildup Slows Down Transcription
rate constant is determined to be 0.78 3 10�3 s�1, comparable
to the pseudo-first-order gyrase-DNA binding rate constant
We examined whether chromosomal supercoiling level affects
�10�3 s�1, which is the product between the bimolecular
transcription elongation in live E. coli cells. Using quantitative
binding rate constant kon z 104 M�1 s�1 under our salt
RT-PCR, we measured the steady-state abundance of different
concentration (and the gyrase
segments of fully induced lac operon mRNA under gyrase
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 319
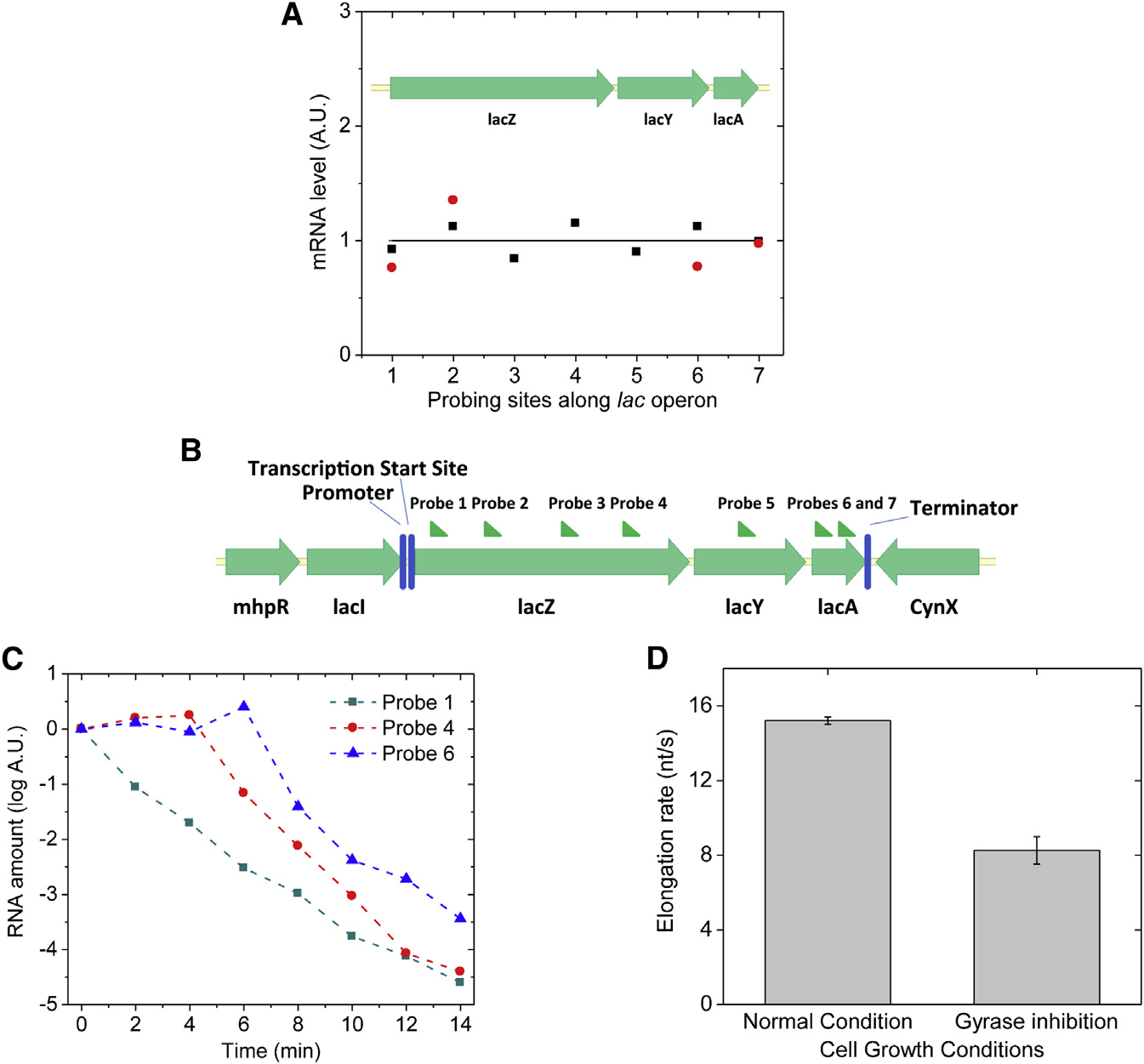
Figure 6. Transcription Processivity andElongation Rate upon Gyrase Inhibition inLive E. coli Cells(A) Quantitative RT-PCR measurement of theabundance of different parts of lac operon mRNAunder fully induced condition. x axis: the positionof probing sites along lac operon; y axis: mRNAabundance. Black squares: gyrase partial inhibi-tion by 50 ng/ml novobiocin; red dots: gyrasecomplete inhibition by 10 ng/ml norfloxacin. Theresult indicates nonstop transcription elongationupon positive supercoiling buildup on the DNA.
The abundance of each mRNA part is normalizedto its abundance under wild-type condition, whichis plotted as the flat curve.
(B) Seven sites on lac operon mRNA that wereprobed in the measurement of transcription elon-gation rate.
(C) Five hundred nanograms per microliter rifam-picin was added into the cell culture at time zero tostop transcription initiation, but not elongation.
The abundance of different positions on the lacoperon mRNA was probed by quantitative RT-PCR at multiple time points.
(D) Transcription elongation rate decreased upongyrase inhibition by 10 ng/ml norfloxacin in liveE. coli cells. The error bars are SDs of the elon-gation rates obtained by repeating the measure-ments (n = 3) under each condition.
See also Figure S6.
inhibition by novobiocin or norfloxacin. No difference in the
viously reported by Higgins group on Salmonella enterica
abundance was observed throughout the transcript A).
This result suggests that the elongation complex does not stopor dissociate from the DNA template in the middle of one round
A Two-State Model Describes Transcriptional Bursting
of transcription more often when the DNA template is more posi-
Transcriptional bursting has been described with a two-state
tively supercoiled. We note that an early in vitro experiment
model, but the origin of the two states was not understood
found that stable norfloxacin-gyrase-DNA complex could form
at a strong gyrase-binding site and block transcription elonga-
We now understand the mechanism of bacterial transcrip-
tion ). This effect was not observed in our
tional bursting A): the gene stochastically switches
live-cell assay, likely due to a low intracellular norfloxacin con-
between on and off states due to release and accumulation
centration and the lack of strong gyrase-binding sites in the
of positive supercoiling. The on state (state 1) generates
probed region.
mRNAs with an average transcription rate k1, and the mRNAs
Next, we measured transcription elongation rate in live E. coli
degrade with rate constant g. The off state (state 2) does not
cells using transcription initiation inhibitor rifampicin (
generate any mRNA. The interconversion rate constants
and quantitative RT-PCR (H. Chen, K. Shiro-
between the two states are a and b. a is the gene on-to-off
guchi, H.G., and X.S.X., unpublished data). We added rifam-
transition rate constant due to gyrase dissociation from
picin to the cell culture at time zero and measured the mRNA
the DNA loop and positive supercoiling accumulation. For
abundance in multiple regions (B) along the transcript
simplicity, we assume positive supercoiling accumulation
at multiple time points afterward. Whereas the mRNA abun-
is fast and gyrase dissociation is rate limiting. b corresponds
dance at the 50 end decreased immediately upon the addition
to the pseudo-first-order rate constant of gyrase-DNA binding,
of rifampicin, the mRNA abundance downstream started to
which is also rate limiting in the gene off-to-on transition
decrease after a time delay C). The distance between
and proportional to the effective intracellular gyrase concen-
the two probes on the transcript divided by the time delay
tration. The lower the effective gyrase concentration is, the
was the elongation rate. Gyrase inhibition was achieved by
longer the gene stays in the off state and the smaller the
norfloxacin treatment where most cells were viable through
on/off duty cycle ratio (b/a), which should result in a higher
the 14-min-long rifampicin assay (Figure S6A). We found that
extent of bursting reflected by a larger Fano factor and a
the elongation rate of fully induced lac operon decreased by
larger fraction of cells that contain zero copy of mRNA at a
46% upon gyrase inhibition similar to the result pre-
given time point.
320 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
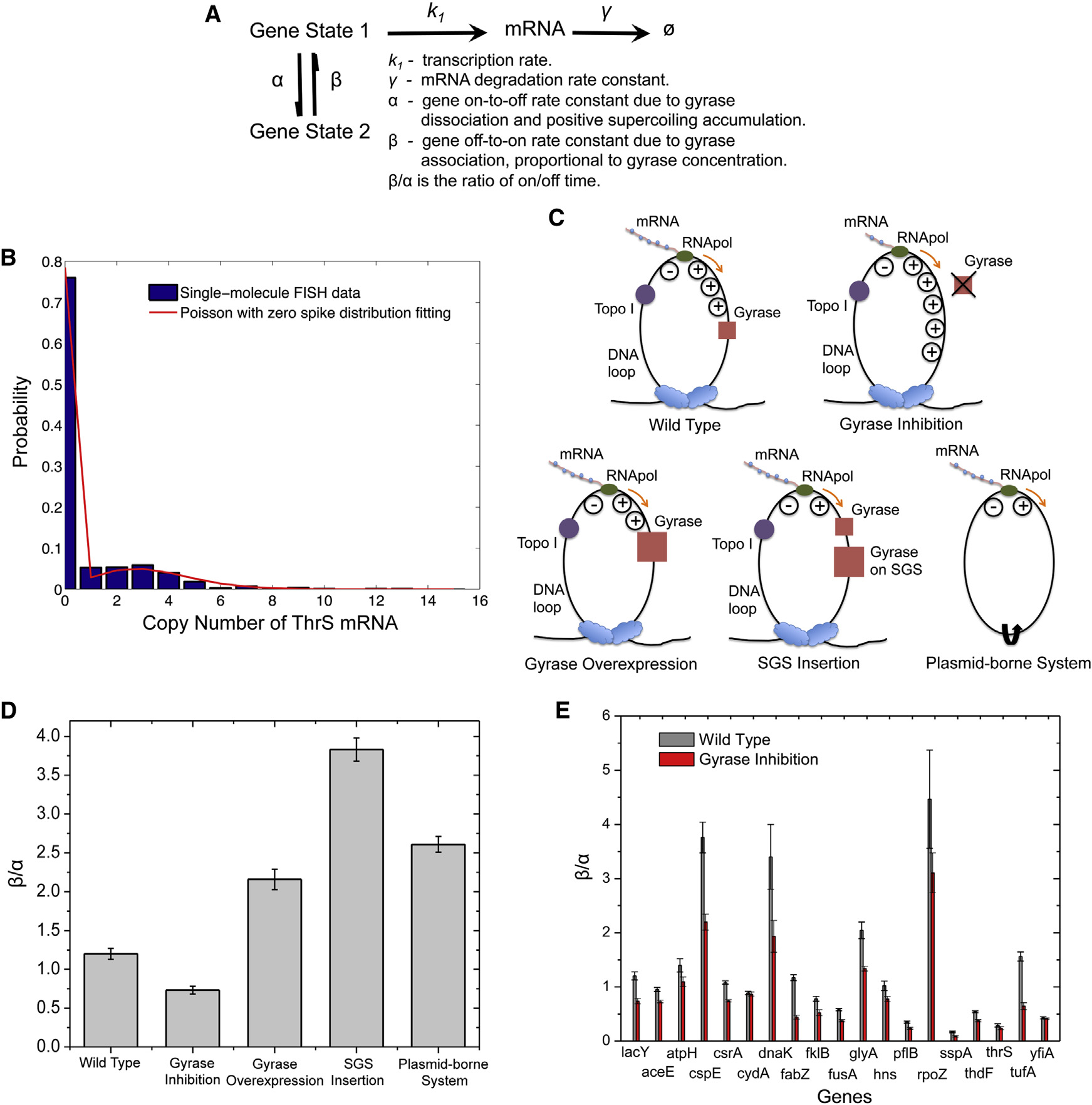
Figure 7. Dependence of On/Off Duty Cycle Ratio, b/a, on Effective Intracellular Gyrase Concentration(A) Kinetic scheme of the two-state model with relevant rate constants.
(B) Fitting of cellular ThrS mRNA copy number distribution with Poisson with zero spike distribution.
(C) Schematics of interactions between effective gyrase concentration and DNA supercoiling generated by transcription under different conditions. Upongyrase inhibition, positive supercoiling accumulates on the chromosomal DNA loop to a higher extent than wild-type. Gyrase overexpression or SGSinsertion is the opposite. In a plasmid-borne expression module, positive and negative supercoiling annihilate each other due to the lack of topologicalbarriers.
(D) b/a of fully induced lac operon decreases upon gyrase partial inhibition by 50 ng/ml novobiocin treatment and increases upon gyrase overexpression, SGSinsertion, and in a plasmid-borne system.
(E) b/a of fully induced lac operon and other 18 highly transcribed E. coli genes. b/a of all the 19 genes decrease upon gyrase partial inhibition by 50 ng/mlnovobiocin treatment. In (D) and (E), the error bars are bootstrapped confidence intervals.
See also Figures S6, S7, and .
Transcription bursts of highly transcribed genes are reflected
tion of cells is bimodal () and can be
by the non-Poissonian mRNA copy number distribution.
approximated with a ‘‘Poisson with zero spike'' distribution
Under the condition that a and b are significantly smaller
(Equation 1). Based on the two-state model, the Fano factor
than k1 and g as previously observed (),
(F) can be derived as Equation 2
the steady-state mRNA copy number distribution for a popula-
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 321
One would expect b/a to be infinitely large if the gene is always
pð0Þ = a +be�k1g + a+b
on in the complete absence of positive supercoiling accumula-
tion. Yet it was not the case for the plasmid-borne system
because there could be weak and transient topological barriers
pðnÞ =
a + b e�k1g
on the plasmid DNA due to transient protein binding (
To confirm this point, we performed control exper-iments on the same plasmid-borne system under gyrase inhibi-
tion and overexpression conditions. We found that b/a changed
in the same direction as the chromosomal gene but to a smallerextent. Under the same conditions, b/a of the plasmid-borne
We note these results hold only under the condition that gyrase
system was always higher than the chromosomal counterpart,
dissociation and rebinding are rate limiting, longer than the
because the plasmid has much lower topological barriers and
time scales of positive supercoiling accumulation and release.
thus more efficient removal of positive supercoiling during active
Although this is a simplified model, it captures the origin of tran-
transcription (Figure S6C).
scriptional bursting, i.e., gyrase dissociation, and establishes
Intriguingly, similar to the scenario of fully induced lac operon,
the gyrase concentration dependence of b, which can now be
all the other 18 genes showed a decreased b/a upon
subject to in vivo experimental tests.
gyrase inhibition. In addition, most of the genes showed anincreased Fano factor (Figure S7A) and an increased fraction
The Dependence of Transcriptional Bursting on
of cells containing zero copy of mRNA (Figure S7B) upon gyrase
Effective Gyrase Concentration Revealed by
inhibition. These findings are consistent with the prediction
Single-Molecule mRNA FISH Assay
based on our model and demonstrate the ubiquitous effect of
We now experimentally verify this model by measuring the
gyrase concentration on transcriptional bursting.
steady-state mRNA copy number distribution in a populationof isogenic E. coli cells under gyrase inhibition and overexpres-
sion conditions. We performed mRNA FISH assay with single-molecule sensitivity, using a single Atto 594-labeled 20-oligomer
Mechanism of Transcriptional Bursting under Induced
nucleotide probing the yfp sequence in an E. coli strain with the
Condition Revealed
target gene fused to yfp sequence endogenously. By measuring
Pertinent to the fact that there is only one (or two) copy of the gene
the intensity of each fluorescent spot and counting the number of
in a cell, gene expression is stochastic. In recent years, stochastic
spots per cell, we determined cellular mRNA copy number for
gene expression has stimulated wide interest
thousands of E. coli cells. The efficiency of our single-molecule
mRNA FISH assay is �90% ).
Such stochasticity, or noise,
We measured the cellular mRNA copy number distribution of
causes phenotypic variability among genetically identical cells
the fully induced lac operon and 18 highly transcribed genes
and organisms despite identical histories of environmental expo-
from the YFP library that our group has constructed
sure ) and arises from the
). Partial gyrase inhibition was achieved by novobiocin
fact that DNA, mRNA, and gene regulatory proteins can be pre-
treatment at low concentration without affecting normal bacterial
sent and active at only a few copies per cell. Due to the small
growth and morphology (Figure S6B). We found that the cellular
copy numbers and the fact that stochastic gene expression
mRNA copy number distribution can be fitted well by the Poisson
cannot be synchronized among different cells, quantitative
with zero spike distribution for the 19 genes with excellent coef-
studies of gene expression at the single-cell level necessitate
ficients of determination ). The fitting allows
single-molecule sensitivity for mRNA and protein detection.
estimation of the on/off duty cycle ratio of transcriptional
The stochastic gene expression dynamics of repressed genes
bursting (b/a), with an error bar obtained by the bootstrapping
have already been well studied and understood to date (
). For highly expressed genes in both prokaryotic and
For fully induced lac operon, b/a was 1.20 for wild-type,
eukaryotic organisms, bursting transcription has been demon-
decreased to 0.73 upon gyrase inhibition, increased to 2.16
strated by a number of techniques, including single-molecule
upon gyrase overexpression, and increased to 3.83 when a
FISH assay that counts cellular mRNA copy number (
strong gyrase site (SGS) was inserted next to the lac operon
C and 7D). This result indicates that transcriptional
PP7 technique that visualizes single mRNA production in real
bursting is sensitively dependent on the availability of gyrase
to remove positive supercoiling accumulated during active
), and fluorescent protein ) or luciferase
If lac operon is on a plasmid that lacks topological constraint,
(as gene expression reporter in live cells.
positive and negative supercoiling generated by transcription
Nevertheless, the mechanism of this ubiquitous phenomenon
could diffuse along the circular DNA in opposite directions and
under induced condition is not understood.
annihilate each other (As a critical control, a
We note that the transcriptional bursting phenomenon studied
plasmid-borne system in E. coli indeed showed an even higher
in this paper is different from transcriptional pausing in prokary-
b/a than that of gyrase overexpression D).
otic and eukaryotic cells (
322 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
which has been studied by recent
single-molecule manipulation (
molecule fluorescence imaging (
) and RNA-sequencing experiments
). Our assay uses
). Whereas pausing de-
RNA staining so that the elongation process on templates with
scribes intermittent elongation of a transcript, bursting describes
any sequence can be easily monitored for multiple rounds of
discontinuous production of many transcripts over a much
transcription on each template. This is a high-throughput mea-
longer time scale and involves inhibition of both transcription
surement because hundreds of templates in one field of view
initiation and elongation.
can be monitored simultaneously. The assay will be generally
We have revealed the origin of stochastic transcriptional
useful for studying other questions in transcription, such as
bursts in bacteria under induced conditions by conducting a
pausing and termination kinetics.
series of in vitro and live-cell experiments and demonstratedthat reversible switching between different chromosomal super-
Other Possible Mechanisms of Transcriptional Bursting
coiling levels via gyrase dissociation from and rebinding to a DNA
The current report proves that stochastic changes of supercoil-
loop gives rise to bursting transcription. We note this is a funda-
ing level in DNA segments due to gyrase dissociation and rebind-
mental mechanism pertinent to the chromosome structure and
ing is a main mechanism that gives rise to bursting transcription
should be applicable to all the highly expressed genes in pro-
of highly expressed genes in bacteria. However, we note there
karyotic cells and even eukaryotic cells. However, the situation
could be other possible causes of bacterial transcriptional
of eukaryotic cells is more complex than that of bacteria due to
bursting, such as the change of chromosomal looping structure
more complicated transcription regulation and the existence of
due to the dissociation and rebinding of nucleoid-associated
nucleosomes ).
proteins, as well as facilitated transcription reinitiation due todynamical gene looping, where an operon DNA places its pro-
A Role of DNA Supercoiling in Gene Expression
moter and terminator in spatial proximity ).
Although they might cause transcription rate fluctuations in addi-
The interaction between DNA supercoiling and gene expression
tion to the supercoiling effect that we observed, none of these
in bacteria has been investigated for decades. Our knowledge
alternative mechanisms have been experimentally proved to
comes primarily from ensemble studies on the relationship be-
switch genes on and off.
tween the global DNA supercoiling level and the gene expressionlevel. On one hand, bacterial DNA supercoiling level affects the
EXPERIMENTAL PROCEDURES
expression of a few E. coli genes called supercoiling-sensitivegenes as well as transcription elongation
In Vitro Single-Molecule Transcription Assay
rate (due to a combined effect of torsional
To measure transcription elongation rates, T7 RNApol (New England Biolabs)
and bending stress sustained by the supercoiled DNA at the
or E. coli RNApol (Epicentre), NTPs, 250 nM SYTO RNASelect, and an oxygenscavenger system were added to transcription buffer. After infusing the mixture
transcription site
into the flow cell containing immobilized DNA templates, a fluorescent movie
On the other hand, local DNA super-
was recorded under 488 nm laser excitation at 0.22 W/cm2. Images were taken
coiling level is generated by transcription, according to the
every 20 s for 60–80 min, and the acquisition time of each image was 5 s.
‘‘twin-domain model'' developed by Wang and Liu groups in
To measure transcription initiation rates, the reaction mixture was the same
as that for elongation rate measurements except that higher concentrations of
RNApol and NTPs were used. The excitation power density was 0.15 W/cm2.
Images were taken every 75 s with 5 s of image acquisition time.
Here, we report a role of DNA supercoilingin gene expression regulation that can only be revealed by sin-
DNA Staining Assay
gle-molecule and single-cell approaches: transient DNA super-
In order to locate the linear and circular templates in the field of view, 100 nM
coiling generated locally during active transcription gives rise
SYTOX Orange (Life Technologies) in 50 mM Tris-HCl buffer (pH 8.0) was used
to transcriptional bursting, which is a major source of gene
to stain the immobilized DNAs after transcription movies were recorded. Fluo-
expression noise that causes cell-to-cell variability in an isogenic
rescent movies were recorded under 532 nm laser excitation with a power
population. Although earlier work proposed DNA supercoiling
density above 4 W/cm2. The image acquisition time was 0.3 s. The imaging
can be involved in bursting transcription
buffer was kept flowing at 8 ml/hr by a syringe pump (PhD 2000; Harvard Appa-ratus) during the movie recording.
we have experimentally proved that supercoilingdynamics is the primary origin of transcriptional bursting.
Single-Molecule mRNA FISH Assay
The BW25993 E. coli cells were grown in M9 medium with 0.4% glycerol,
In Vitro, Single-Molecule Transcription Assay
amino acids, and vitamins, together with antibiotics and saturating amount
In order to investigate the effect of positive supercoiling buildup
of isopropyl b-D-thiogalactopyranoside (IPTG) if necessary. The cells were
on transcription elongation and initiation in a clean and con-
subsequently inoculated 1:500 into the same medium and incubated for
trolled fashion, we developed an in vitro, single-molecule assay
�7 hr at 37�C with 250 rpm shaking till optical density 600 nm (OD600nm)reached 0.2�0.3. Fifty nanograms per microliter novobiocin (Sigma) was
that could monitor real-time transcription on individual DNA
added into the medium and incubated for another 2 hr before harvest. Two
templates. We note our assay is different from other existing
hours was long enough (several cell cycles) to allow all the cells to enter steady
in vitro transcription assays using single-molecule manipulation
state and thus minimized potential cell-to-cell variation due to different transi-
tion kinetics in response to the drug treatment.
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 323
The YFP library strains were grown in Luria broth (LB) medium with chloram-
Efron, B., and Tibshirani, R.J. (1993). An Introduction to the Bootstrap (New
phenicol at 30�C. The cells were subsequently inoculated 1:400 into M9
York: Chapman & Hall).
medium with 0.4% glucose, amino acids, and vitamins and incubated for
Bustamante, C., Cheng, W., and Mejia, Y.X. (2011). Revisiting the central
11 hr at 30�C with 250 rpm shaking till OD600nm reached 0.2�0.3. Fifty nano-
dogma one molecule at a time. Cell 144, 480–497.
grams per microliter novobiocin was added and incubated for another 2 hr
Chakraborty, A., Wang, D., Ebright, Y.W., Korlann, Y., Kortkhonjia, E., Kim, T.,
before harvest.
Chowdhury, S., Wigneshweraraj, S., Irschik, H., Jansen, R., et al. (2012). Open-
Single-molecule mRNA FISH assay was performed as previously described
ing and closing of the bacterial RNA polymerase clamp. Science 337, 591–595.
using Venus495r mRNA FISH probe covalently linked to
Cheng, B., Zhu, C.X., Ji, C., Ahumada, A., and Tse-Dinh, Y.C. (2003). Direct
a dye molecule Atto 594 (Sigma-Aldrich). The images were taken under epi-
interaction between Escherichia coli RNA polymerase and the zinc ribbon do-
illumination by a fiber laser at 580 nm and phase contrast illumination by a
mains of DNA topoisomerase I. J. Biol. Chem.
halogen lamp.
Choi, P.J., Cai, L., Frieda, K., and Xie, X.S. (2008). A stochastic single-moleculeevent triggers phenotype switching of a bacterial cell. Science
SUPPLEMENTAL INFORMATION
Chubb, J.R., Trcek, T., Shenoy, S.M., and Singer, R.H. (2006). Transcriptional
Supplemental Information includes Extended Experimental Procedures, seven
pulsing of a developmental gene. Curr. Biol. 16, 1018–1025.
figures, and one table and can be found with this article online at
Churchman, L.S., and Weissman, J.S. (2011). Nascent transcript sequencing
visualizes transcription at nucleotide resolution. Nature 469, 368–373.
Core, L.J., Waterfall, J.J., and Lis, J.T. (2008). Nascent RNA sequencing
AUTHOR CONTRIBUTIONS
reveals widespread pausing and divergent initiation at human promoters.
Science 322, 1845–1848.
X.S.X. conceived the project and supervised the experiments. S.C. devel-
Davenport, R.J., Wuite, G.J., Landick, R., and Bustamante, C. (2000). Single-
oped the in vitro, single-molecule transcription assay. S.C. performed the
molecule study of transcriptional pausing and arrest by E. coli RNA polymer-
in vitro imaging experiments, data analysis, and biophysical calculations
ase. Science 287, 2497–2500.
based on the in vitro data. S.C. and C.C. performed the control of theenzyme activities in the in vitro, single-molecule transcription assay. C.C.
Deng, S., Stein, R.A., and Higgins, N.P. (2004). Transcription-induced barriers
performed the single-molecule mRNA FISH assay, data analysis, and live-
to supercoil diffusion in the Salmonella typhimurium chromosome. Proc. Natl.
cell experiments based on quantitative RT-PCR. C.C. made the DNA con-
Acad. Sci. USA 101, 3398–3403.
structs for the in vitro, single-molecule assay and the FISH assay. H.G. built
Deng, S., Stein, R.A., and Higgins, N.P. (2005). Organization of supercoil do-
the mathematical model. H.G., C.C., and S.C fitted the model to the single-
mains and their reorganization by transcription. Mol. Microbiol. 57, 1511–1521.
molecule FISH data. S.C., C.C., and X.S.X. designed the experiments and
Drlica, K. (1992). Control of bacterial DNA supercoiling. Mol. Microbiol. 6,
wrote the manuscript.
Drolet, M. (2006). Growth inhibition mediated by excess negative supercoiling:
the interplay between transcription elongation, R-loop formation and DNAtopology. Mol. Microbiol. 59, 723–730.
We thank Xiaowei Zhuang for the collaboration on bacterial chromosomal
El Hanafi, D., and Bossi, L. (2000). Activation and silencing of leu-500 promoter
structure study, which prompted us to conduct the current study; N. Patrick
by transcription-induced DNA supercoiling in the Salmonella chromosome.
Higgins for providing the plasmid containing strong gyrase site sequence;
Mol. Microbiol. 37, 583–594.
Gene-Wei Li for development of FISH protocol; Minbiao Ji for help with micro-scope construction; Rahul Roy for advice on data analysis; and James Wang,
Elowitz, M.B., and Leibler, S. (2000). A synthetic oscillatory network of tran-
Long Cai, Gene-Wei Li, and Paul Choi for critical reading of the manuscript.
scriptional regulators. Nature 403, 335–338.
This work was supported by NIH Pioneer Award (1DP1OD000277; to X.S.X.),
Elowitz, M.B., Levine, A.J., Siggia, E.D., and Swain, P.S. (2002). Stochastic
NIH grant TR01 (5R01GM096450-02; to X.S.X.), National Science Foundation
gene expression in a single cell. Science 297, 1183–1186.
of China (21373021; to H.G.), and the Foundation for the Author of National
Epshtein, V., and Nudler, E. (2003). Cooperation between RNA polymerase
Excellent Doctoral Dissertation of China (201119; to H.G.).
molecules in transcription elongation. Science 300, 801–805.
Franco, R.J., and Drlica, K. (1988). DNA gyrase on the bacterial chromosome.
Received: September 25, 2013
Oxolinic acid-induced DNA cleavage in the dnaA-gyrB region. J. Mol. Biol.
Revised: March 17, 2014
Accepted: May 8, 2014
Friedman, L.J., and Gelles, J. (2012). Mechanism of transcription initiation at
Published: July 17, 2014
an activator-dependent promoter defined by single-molecule observation.
Cell 148, 679–689.
Golding, I., Paulsson, J., Zawilski, S.M., and Cox, E.C. (2005). Real-time
Abbondanzieri, E.A., Greenleaf, W.J., Shaevitz, J.W., Landick, R., and Block,
kinetics of gene activity in individual bacteria. Cell 123, 1025–1036.
S.M. (2005). Direct observation of base-pair stepping by RNA polymerase.
Gore, J., Bryant, Z., Stone, M.D., No¨llmann, M., Cozzarelli, N.R., and Busta-
Nature 438, 460–465.
mante, C. (2006). Mechanochemical analysis of DNA gyrase using rotor
Bai, L., Santangelo, T.J., and Wang, M.D. (2006). Single-molecule analysis of
bead tracking. Nature 439, 100–104.
RNA polymerase transcription. Annu. Rev. Biophys. Biomol. Struct. 35,
Guptasarma, P. (1996). Cooperative relaxation of supercoils and periodic tran-
scriptional initiation within polymerase batteries. BioEssays 18, 325–332.
Baker, T.A., Funnell, B.E., and Kornberg, A. (1987). Helicase action of dnaB
Hardy, C.D., and Cozzarelli, N.R. (2005). A genetic selection for supercoiling
protein during replication from the Escherichia coli chromosomal origin
mutants of Escherichia coli reveals proteins implicated in chromosome struc-
in vitro. J. Biol. Chem. 262, 6877–6885.
ture. Mol. Microbiol. 57, 1636–1652.
Billingsley, D.J., Bonass, W.A., Crampton, N., Kirkham, J., and Thomson, N.H.
Hebenstreit, D. (2013). Are gene loops the cause of transcriptional noise?
(2012). Single-molecule studies of DNA transcription using atomic force micro-
Trends Genet. 29, 333–338.
scopy. Phys. Biol. 9, 021001.
Herbert, K.M., La Porta, A., Wong, B.J., Mooney, R.A., Neuman, K.C., Landick,
Blake, W.J., KAErn, M., Cantor, C.R., and Collins, J.J. (2003). Noise in eukary-
R., and Block, S.M. (2006). Sequence-resolved detection of pausing by single
otic gene expression. Nature 422, 633–637.
RNA polymerase molecules. Cell 125, 1083–1094.
324 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
Herbert, K.M., Greenleaf, W.J., and Block, S.M. (2008). Single-molecule
Munsky, B., Neuert, G., and van Oudenaarden, A. (2012). Using gene expres-
studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 77,
sion noise to understand gene regulation. Science 336, 183–187.
Muramoto, T., Cannon, D., Gierlinski, M., Corrigan, A., Barton, G.J., and
Higgins, N.P., and Cozzarelli, N.R. (1982). The binding of gyrase to DNA: anal-
Chubb, J.R. (2012). Live imaging of nascent RNA dynamics reveals distinct
ysis by retention by nitrocellulose filters. Nucleic Acids Res. 10, 6833–6847.
types of transcriptional pulse regulation. Proc. Natl. Acad. Sci. USA 109,
Higgins, N.P., Peebles, C.L., Sugino, A., and Cozzarelli, N.R. (1978). Purifica-
tion of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic
Ozbudak, E.M., Thattai, M., Kurtser, I., Grossman, A.D., and van Oudenaar-
activity. Proc. Natl. Acad. Sci. USA 75, 1773–1777.
den, A. (2002). Regulation of noise in the expression of a single gene. Nat.
Hocine, S., Raymond, P., Zenklusen, D., Chao, J.A., and Singer, R.H. (2013).
Genet. 31, 69–73.
Single-molecule analysis of gene expression using two-color RNA labeling in
Pedraza, J.M., and Paulsson, J. (2008). Effects of molecular memory and
live yeast. Nat. Methods 10, 119–121.
bursting on fluctuations in gene expression. Science 319, 339–343.
Hodges, C., Bintu, L., Lubkowska, L., Kashlev, M., and Bustamante, C. (2009).
Peter, B.J., Arsuaga, J., Breier, A.M., Khodursky, A.B., Brown, P.O., and Coz-
Nucleosomal fluctuations govern the transcription dynamics of RNA polymer-
zarelli, N.R. (2004). Genomic transcriptional response to loss of chromosomal
ase II. Science 325, 626–628.
supercoiling in Escherichia coli. Genome Biol. 5, R87.
Kannemeier, C., Shibamiya, A., Nakazawa, F., Trusheim, H., Ruppert, C., Mar-
Postow, L., Hardy, C.D., Arsuaga, J., and Cozzarelli, N.R. (2004). Topological
kart, P., Song, Y., Tzima, E., Kennerknecht, E., Niepmann, M., et al. (2007).
domain structure of the Escherichia coli chromosome. Genes Dev. 18, 1766–
Extracellular RNA constitutes a natural procoagulant cofactor in blood coagu-
lation. Proc. Natl. Acad. Sci. USA 104, 6388–6393.
Raj, A., Peskin, C.S., Tranchina, D., Vargas, D.Y., and Tyagi, S. (2006).
Kapanidis, A.N., Margeat, E., Ho, S.O., Kortkhonjia, E., Weiss, S., and Ebright,
Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309.
R.H. (2006). Initial transcription by RNA polymerase proceeds through a DNA-
Raj, A., Rifkin, S.A., Andersen, E., and van Oudenaarden, A. (2010). Variability
scrunching mechanism. Science 314, 1144–1147.
in gene expression underlies incomplete penetrance. Nature 463, 913–918.
Kussell, E., and Leibler, S. (2005). Phenotypic diversity, population growth, and
Raser, J.M., and O'Shea, E.K. (2004). Control of stochasticity in eukaryotic
information in fluctuating environments. Science 309, 2075–2078.
gene expression. Science 304, 1811–1814.
Landick, R. (2006). The regulatory roles and mechanism of transcriptional
Rau, D.C., Gellert, M., Thoma, F., and Maxwell, A. (1987). Structure of the DNA
pausing. Biochem. Soc. Trans. 34, 1062–1066.
gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol.
Larson, D.R., Zenklusen, D., Wu, B., Chao, J.A., and Singer, R.H. (2011). Real-
time observation of transcription initiation and elongation on an endogenous
Reece, R.J., and Maxwell, A. (1991). DNA gyrase: structure and function. Crit.
yeast gene. Science 332, 475–478.
Rev. Biochem. Mol. Biol. 26, 335–375.
Leng, F., Chen, B., and Dunlap, D.D. (2011). Dividing a supercoiled DNA mole-
Revyakin, A., Ebright, R.H., and Strick, T.R. (2004). Promoter unwinding and
cule into two independent topological domains. Proc. Natl. Acad. Sci. USA
promoter clearance by RNA polymerase: detection by single-molecule DNA
nanomanipulation. Proc. Natl. Acad. Sci. USA 101, 4776–4780.
Li, G.W., and Xie, X.S. (2011). Central dogma at the single-molecule level in
Revyakin, A., Zhang, Z., Coleman, R.A., Li, Y., Inouye, C., Lucas, J.K., Park,
living cells. Nature 475, 308–315.
S.R., Chu, S., and Tjian, R. (2012). Transcription initiation by human RNA
Lim, H.M., Lewis, D.E., Lee, H.J., Liu, M., and Adhya, S. (2003). Effect of vary-
polymerase II visualized at single-molecule resolution. Genes Dev. 26, 1691–
ing the supercoiling of DNA on transcription and its regulation. Biochemistry
Rovinskiy, N., Agbleke, A.A., Chesnokova, O., Pang, Z., and Higgins, N.P.
Lionberger, T.A., and Meyho¨fer, E. (2010). Bending the rules of transcrip-
(2012). Rates of gyrase supercoiling and transcription elongation control
tional repression: tightly looped DNA directly represses T7 RNA polymerase.
supercoil density in a bacterial chromosome. PLoS Genet. 8, e1002845.
Biophys. J. 99, 1139–1148.
Samul, R., and Leng, F. (2007). Transcription-coupled hypernegative super-
Lionnet, T., Czaplinski, K., Darzacq, X., Shav-Tal, Y., Wells, A.L., Chao, J.A.,
coiling of plasmid DNA by T7 RNA polymerase in Escherichia coli topoisomer-
Park, H.Y., de Turris, V., Lopez-Jones, M., and Singer, R.H. (2011). A trans-
ase I-deficient strains. J. Mol. Biol. 374, 925–935.
genic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods
Shundrovsky, A., Santangelo, T.J., Roberts, J.W., and Wang, M.D. (2004).
A single-molecule technique to study sequence-dependent transcription
Liu, L.F., and Wang, J.C. (1987). Supercoiling of the DNA template during tran-
pausing. Biophys. J. 87, 3945–3953.
scription. Proc. Natl. Acad. Sci. USA 84, 7024–7027.
Singh, A., Razooky, B., Cox, C.D., Simpson, M.L., and Weinberger, L.S. (2010).
Lynch, A.S., and Wang, J.C. (1993). Anchoring of DNA to the bacterial cyto-
Transcriptional bursting from the HIV-1 promoter is a significant source of sto-
plasmic membrane through cotranscriptional synthesis of polypeptides en-
chastic noise in HIV-1 gene expression. Biophys. J. 98, L32–L34.
coding membrane proteins or proteins for export: a mechanism of plasmid
Skinner, G.M., Baumann, C.G., Quinn, D.M., Molloy, J.E., and Hoggett, J.G.
hypernegative supercoiling in mutants deficient in DNA topoisomerase I.
(2004). Promoter binding, initiation, and elongation by bacteriophage T7
J. Bacteriol. 175, 1645–1655.
RNA polymerase. A single-molecule view of the transcription cycle. J. Biol.
Ma, J., Bai, L., and Wang, M.D. (2013). Transcription under torsion. Science
Chem. 279, 3239–3244.
Snyder, M., and Drlica, K. (1979). DNA gyrase on the bacterial chromosome:
Maamar, H., Raj, A., and Dubnau, D. (2007). Noise in gene expression deter-
DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131, 287–302.
mines cell fate in Bacillus subtilis. Science 317, 526–529.
So, L.H., Ghosh, A., Zong, C., Sepu´lveda, L.A., Segev, R., and Golding, I.
Maxwell, A., and Gellert, M. (1984). The DNA dependence of the ATPase activ-
(2011). General properties of transcriptional time series in Escherichia coli.
ity of DNA gyrase. J. Biol. Chem. 259, 14472–14480.
Nat. Genet. 43, 554–560.
Mitarai, N., Dodd, I.B., Crooks, M.T., and Sneppen, K. (2008). The generation
Suter, D.M., Molina, N., Gatfield, D., Schneider, K., Schibler, U., and Naef, F.
of promoter-mediated transcriptional noise in bacteria. PLoS Comput. Biol. 4,
(2011). Mammalian genes are transcribed with widely different bursting
kinetics. Science 332, 472–474.
Morrison, A., and Cozzarelli, N.R. (1981). Contacts between DNA gyrase and
Tang, G.Q., Roy, R., Bandwar, R.P., Ha, T., and Patel, S.S. (2009). Real-time
its binding site on DNA: features of symmetry and asymmetry revealed by pro-
observation of the transition from transcription initiation to elongation of the
tection from nucleases. Proc. Natl. Acad. Sci. USA 78, 1416–1420.
RNA polymerase. Proc. Natl. Acad. Sci. USA 106, 22175–22180.
Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc. 325
Taniguchi, Y., Choi, P.J., Li, G.W., Chen, H., Babu, M., Hearn, J., Emili, A., and
Willmott, C.J., Critchlow, S.E., Eperon, I.C., and Maxwell, A. (1994). The com-
Xie, X.S. (2010). Quantifying E. coli proteome and transcriptome with single-
plex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcrip-
molecule sensitivity in single cells. Science 329, 533–538.
tion by RNA polymerase. J. Mol. Biol. 242, 351–363.
ten Heggeler-Bordier, B., Wahli, W., Adrian, M., Stasiak, A., and Dubochet, J.
Wolf, D.M., Vazirani, V.V., and Arkin, A.P. (2005). Diversity in times of adversity:
(1992). The apical localization of transcribing RNA polymerases on supercoiled
probabilistic strategies in microbial survival games. J. Theor. Biol. 234,
DNA prevents their rotation around the template. EMBO J. 11, 667–672.
Thattai, M., and van Oudenaarden, A. (2004). Stochastic gene expression in
Wu, H.Y., Shyy, S.H., Wang, J.C., and Liu, L.F. (1988). Transcription generates
fluctuating environments. Genetics 167, 523–530.
positively and negatively supercoiled domains in the template. Cell 53,
Tsao, Y.P., Wu, H.Y., and Liu, L.F. (1989). Transcription-driven supercoiling of
DNA: direct biochemical evidence from in vitro studies. Cell 56, 111–118.
Zhang, Z., Revyakin, A., Grimm, J.B., Lavis, L.D., and Tjian, R. (2014). Single-
Wang, W., Li, G.W., Chen, C., Xie, X.S., and Zhuang, X. (2011). Chromosome
molecule tracking of the transcription cycle by sub-second RNA detection.
organization by a nucleoid-associated protein in live bacteria. Science 333,
eLife 3, e01775.
Zong, C., So, L.H., Sepu´lveda, L.A., Skinner, S.O., and Golding, I. (2010).
Weixlbaumer, A., Leon, K., Landick, R., and Darst, S.A. (2013). Structural basis
Lysogen stability is determined by the frequency of activity bursts from the
of transcriptional pausing in bacteria. Cell 152, 431–441.
fate-determining gene. Mol. Syst. Biol. 6, 440.
326 Cell 158, 314–326, July 17, 2014 ª2014 Elsevier Inc.
Source: https://bernstein.harvard.edu/papers/1-s2.0-S0092867414007399-main.pdf
ISSN 0943-6839 18 Euro / 32 CHF Die Säuren in der Homöopathie Rund um die Geburt DIE HZ II/2016 ERSCHEINT IM JULI 2016 und um die GR Autoren:Gabriele Bengler Peter Bergmann Micha Bitschnau Marijke Creveld Monika Grasser Rotger Heilmeier ATHIE ZEITSCHRIFT Eva Kolbinger Christine Lauterbach Monika Liewers Angela Orendt-Stolp Anne Schadde Gabriele Schubert
An interprofessional case study THIS CLINICAL TRAINING INITIATIVE IS SUPPORTED BY FUNDING FROM THE AUSTRALIAN GOVERNMENT UNDER THE INCREASED CLINICAL TRAINING CAPACITY (ICTC) PROGRAM Asthma - an interprofessional case study At the end of this presentation students will be able to: • Give a definition of asthma• Discuss what could cause asthma• Identify four triggers of asthma• Describe symptoms of an asthma flare-up• Explain three asthma treatment strategies• Develop an interprofessional plan of care for















