X96411u06
Cell, Vol. 103, 853–864, December 8, 2000, Copyright
2000 by Cell Press
Synergism with the Coactivator OBF-1
(OCA-B, BOB-1) Is Mediated by a Specific
POU Dimer Configuration
Alexey Tomilin,*†# Attila Reme´nyi,†§#
responsible for regulating the expression of develop-
Katharina Lins,* Hanne Bak,† Sebastian Leidel,*
mental genes. This diversity in transcriptional control
Gerrit Vriend,‡ Matthias Wilmanns,§
by a limited array of transcription factors is achieved
and Hans R. Scho¨ler*†k
through a complex network of interactions between
*Center for Animal Transgenesis
these proteins and specific DNA sequences found in
and Germ Cells Research
promoters and enhancers of developmental genes. The
New Bolton Center
primary structure of these DNA elements defines the
School of Veterinary Medicine
composition and architecture of the transcriptional acti-
Department of Animal Biology
vation complexes that ultimately control gene expres-
University of Pennsylvania
sion in the appropriate temporo-spatial context of the
Kennett Square, Pennsylvania 19348
developing organism. For example, nonsteroid mem-
†Gene Expression Programme
bers of the nuclear receptor superfamily that possess
a zinc-finger DNA binding domain operate by binding
69117 Heidelberg, Germany
to the hormone response elements (HREs). HREs con-
sist of two minimal core hexad sequences, AGGTCA,
which can be configured into various functional motifs.
The orientation and spacing between these two hexa-
69117 Heidelberg, Germany
§
mers as well as subtle differences in their sequence
dictate the identity and the mode (monomer, hetero-, or
EMBL, c/o DESY
homodimer) of nuclear receptor binding that results in
Notkestr 85
diverse effects on transcription (Mangelsdorf and Evans,
2263 Hamburg, Germany
The operation of members of the POU domain family
of transcription factors is also highly dependent on the
nature of cognate DNA elements. The 160 amino-acid-
long DNA binding domain of these proteins is composed
of two structurally independent subdomains: the POU-
POU domain proteins contain a bipartite DNA binding
type homeodomain (POU-homeo or POUH), and the
domain divided by a flexible linker that enables them
POU-specific domain (POUS) that are connected by a
to adopt various monomer configurations on DNA. The
flexible linker region (Scho¨ler, 1991; Verrijzer and van
versatility of POU protein operation is additionally con-
der Vliet, 1993). POU domain proteins demonstrate im-
ferred at the dimerization level. The POU dimer formed
pressive versatility in how they regulate transcription.
on the PORE (ATTTGAAATGCAAAT) can recruit the
This is due to several, often interdependent, factors: (1)
transcriptional coactivator OBF-1, whereas POU di-
flexible amino acid–base interaction, (2) variable orienta-
mers formed on the consensus MORE (ATGCATATG
tion, spacing, and positioning of DNA-tethered POU
CAT) or on MOREs from immunoglobulin heavy chain
subdomains relative to each other, (3) posttranslational
promoters (AT[G/A][C/A]ATATGCAA) fail to interact.
modification, and (4) interaction with heterologous pro-
An interaction with OBF-1 is precluded since the same
teins (Herr and Cleary, 1995).
Oct-1 residues that form the MORE dimerization inter-
POU domain proteins are able to bind to DNA cooper-
face are also used for OBF-1/Oct-1 interactions on the
atively, thus conferring additional functional variability.
PORE. Our findings provide a paradigm of how specific
The homo- and heterodimerization of Oct-1 and Oct-2
POU dimer assemblies can differentially recruit a coreg-
on immunoglobulin (Ig) heavy chain promoters (VH) pro-
ulatory activity with distinct transcriptional readouts.
vided evidence of cooperativity, with a yet unknown
dimer arrangement (Kemler et al., 1989; LeBowitz et
al., 1989; Poellinger et al., 1989). The cis-elements are
considered to consist of low-affinity heptamer and high-
affinity octamer sites separated by two nucleotides
Development of multicellular organisms is characterized
by an intricate series of genetic and epigenetic events
The pituitary-specific POU domain protein Pit-1 binds
that generate the complex adult body from the unicellu-
to DNA either as a homodimer or as a heterodimer with
lar zygote. A refined and sophisticated regulatory net-
Oct-1 (Voss et al., 1991). Crystallographic studies deter-
work that is established during embryogenesis reflects
mined the structure of a Pit-1 homodimer assembled
the complexity of organisms. Although embryonic devel-
on the synthetic motif ATGTATATACAT (referred to here
opment is a multistep process characterized by the se-
as PitD) that had been derived from the natural Pit-1
quential activation and repression of many genes, only
cognate element within the prolactin gene promoter
a relatively small number of transcription factors are
(ATATATATTCAT) (Jacobson et al., 1997). The structure
of the Pit-1 POUS and POUH domains, and their docking
onto DNA, are very similar to that observed in the cocrys-
To whom correspondence should be addressed (e-mail: scholer@
tal of the Oct-1 POU domain monomer with the octamer
# These authors contributed equally to this work.
site (ATGCAAAT, Klemm et al., 1994). The Oct-1 POUS
domain recognizes the ATGC subsite whereas the Pit-1
two Oct factors (Figure 1B). The identity of these com-
POUS domain binds to the sequence ATAC. However,
plexes was subsequently confirmed using Oct-1 and
the latter subsite lies on the opposite strand and, as a
Oct-4 antibodies (Figure 1C). Further in vitro analyses
consequence, the orientation of POUS relative to the
demonstrated that Oct-2 and Oct-6 could also bind to
POUH domain is inverted (Jacobson et al., 1997).
the MORE as homodimers (Figure 2).
Another mechanism outlining cooperative DNA bind-
The Oct proteins studied in this work have overlapping
ing by POU proteins was recently determined during the
temporo-spatial expression patterns during embryo-
course of an Oct-4 target gene characterization (Botquin
genesis and in adult tissues (Scho¨ler et al., 1989;
et al., 1998). The Palindromic Oct factor Recognition
Scho¨ler, 1991; Herr and Cleary, 1995; Ryan and Rosen-
Element (PORE), ATTTGAAATGCAAAT (15 bp), of the
feld, 1997). Oct-1 is coexpressed with Oct-2 (lymphoid
Osteopontin (OPN) enhancer interacts with an Oct-4 di-
cells and some cell types of central nervous system)
mer, thereby mediating strong transcriptional activation
and with Oct-4 and Oct-6 (embryonic pluripotent cells).
in preimplantation mouse embryos. Homo- and hetero-
Subsequently, Oct-1/Oct-2, Oct-1/Oct-4, Oct-1/Oct-6,
dimerization of other Oct factors like Oct-1 and Oct-6
and Oct-4/Oct-6 heterodimers can be formed on the
on the PORE has also been demonstrated.
MORE (Figure 2). Thus, the ability to heterodimerize on
The aforementioned examples provide evidence of
the MORE is a property shared by all four tested Oct
the various ways in which POU domain proteins are able
to cooperatively bind to substrate DNA. The particular
mode of binding employed is primarily defined by the
The MORE and PORE Mediate Different Domain
DNA sequence. To address the question of whether
Arrangements of POU Factor Dimers
diversity in cooperative binding is reflected in transcrip-
The MORE arrangement is depicted schematically in
tional regulation, we have assessed and compared the
Figure 3A (left) on the basis of a crystal structure of
ability of two different types of POU dimers to interact
the Oct-1 POU domain bound to the MORE as a dimer
with the coactivator OBF-1 (OCA-B, Bob-1). This coacti-
recently solved at 1.9 A
˚ resolution (A. R. et al., unpub-
vator synergistically interacts with Oct-1 and Oct-2
lished). The study revealed that the POUS domains inter-
monomers bound to the octamer motif (Luo et al., 1992;
act mainly with the ATGC sequences of the palindromic
Gstaiger et al., 1995; Luo and Roeder, 1995; Strubin et
half-sites of the MORE, whereas the POUH domains bind
al., 1995). We have investigated one type of POU dimer
to the AT sequences. The dimer interface between the
that is formed on the PORE and another that is formed
two POU molecules is formed by a loop region within the
on another palindromic DNA motif called MORE (More
POUS domain (between helices 3 and 4) of one molecule
PORE), ATGCATATGCAT. The data presented in this
interacting with the C-terminal end of the recognition
study provide an example of how POU domain mole-
helix of the POUH of a second molecule. The arrange-
cules that bind to DNA in the same stoichiometry but
ment of the POU subdomains is that reported for the
in different configurations can differentially recruit a
Pit-1/PitD cocrystal (Figure 3A, left; Jacobson et al.,
transcriptional coactivator to the promoter resulting in
1997), the arrangement of the POU:PORE dimeric com-
plex is based on mutagenesis analysis and computer
modeling (Figure 3A, right; Botquin et al., 1998).
In the MORE complex, one half-site binds POUS and
POUH domains from two different Oct molecules. In the
Dimerization of Oct Factors on the MORE
PORE model, the two subdomains bound to one half-
We asked whether the DNA binding configuration exem-
site originate from the same protein molecule. The POU
plified by the Pit-1:PitD crystal structure (Jacobson et
dimers assembled on the PORE and MORE should be-
al., 1997) is a conserved property of the POU family. We
have differentially in terms of their tolerance to base-
focused our analysis on POU proteins that had been
pair insertions between the half-sites. Insertions in the
identified by virtue of their binding to the octamer motif,
center of the PORE alter the relative positions of POU
previously termed Oct factors (Scho¨ler et al., 1989;
domains belonging to two different POU molecules (Bot-
quin et al., 1998). This is an indication that the altered
In an electrophoretic mobility shift assay (EMSA), both
arrangement on the DNA abolishes dimer formation due
naturally and bacterially expressed Oct-1 and Oct-4
to a loss of interface contacts. In contrast, extra base-
formed monomers and dimers on the PitD site (Figure
pair insertions in the center of the MORE (Figure 3B)
1B). We anticipated however that this site was not the
should not affect dimerization. The length of the flexible
ideal substrate for cooperative binding of these Oct fac-
linker connecting the POUS and POUH domains within
tors. Indeed, in the Pit-1:PitD cocrystal, Pit-1 POU
each monomer, however, would impose one possible
mains are docked onto two ATAC subsites on both DNA
limitation to the extent of spacing. In agreement with
faces (Jacobson et al., 1997 and Figure 1A). The POU
this notion, Oct-4 dimerization can tolerate up to two
domain of Oct-1 selects ATGC from a pool of random
base-pair insertions between the half-sites in the MORE
sequences (Verrijzer et al., 1992) and provides all base
(Figure 3C).
contacts within the same sequence in the Oct-1:octamer
crystal (Klemm et al., 1994). Based on this, we converted
The MORE Can Mediate Transcriptional
the two POUS docking subsites within PitD (Figure 1A).
Activation by Oct Factors
Compared to the original motif, the resulted sequence
Next, the MORE was compared to the PORE for the
(MORE) mediated assembly of more stable Oct-1 and
ability to mediate transcriptional activation by Oct factor
Oct-4 homodimers and a heterodimer between these
in transient transfection assays. The PORE was used as
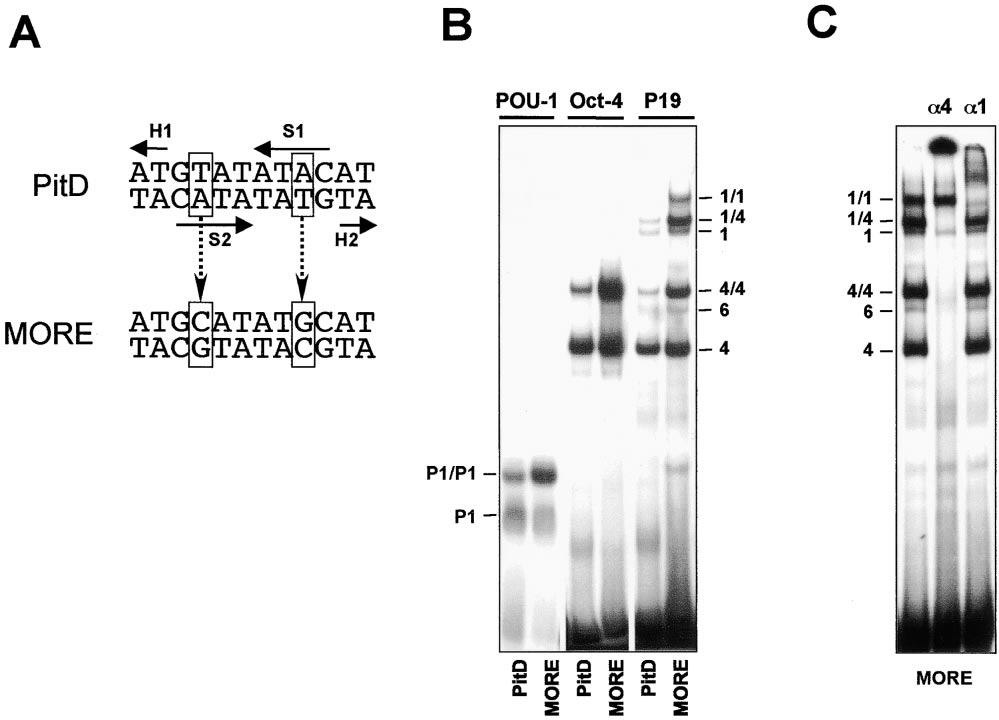
OBF-1 Coactivator Synergy Is Mediated by POU Dimer
855
Figure 1. Two Replacements within a Pit-1 Dimer Binding Site Increase Cooperative Binding of Oct Factor
(A) Sequences of PitD motif and the MORE. Solid arrows indicate the relative orientation (from N to C terminus) and the positioning of two
POUS (S) and two POUH (H) subdomains of Pit-1 (Jacobson et al., 1997). The MORE was derived from the PitD site by replacing the base pairs
shown in boxes.
(B) An EMSA performed to compare the ability of PitD- and MORE-containing oligonucleotides to bind to the bacterially expressed POU
domain of Oct-1 (POU-1), recombinant Oct-4 (Oct-4), and natural Oct-1 and Oct-4 proteins present in a whole-cell extract of P19 embryonic
carcinoma (EC) cells.
(C) EMSA of P19 cell extracts using the MORE oligonucleotide as a probe. Anti-Oct-4 (␣4) or anti-Oct-1 (␣1) antibodies were included in the
binding reaction before applying it onto the gel to prove the identity of the complexes. The ␣1 had only limited effect on Oct-1-containing
complexes, which may have been due to a low affinity of this antibody. The protein–DNA complexes are denoted as follows: P1 and P1/P1,
bacterial POU-1 monomer and homodimer, respectively; 1 and 1/1, natural Oct-1 monomer and homodimer, respectively; 6, Oct-6 monomer;
4 and 4/4, monomer and homodimer of both recombinant and natural Oct-4; and 1/4, Oct-1/Oct-4 heterodimer.
a reference, because it had been shown to be highly
pressed in 293 cells. The ability of Oct-4 to efficiently
active in EC cells (Botquin et al., 1998). Transient trans-
activate transcription via the MOREs and POREs may
fection of the reporters into P19 EC cells demonstrates
be due to the presence of E1A protein in 293 cells,
that the MORE can mediate transcriptional activation
a transcriptional coactivator that can enhance Oct-4
as efficiently as the PORE (Figure 4A). The Oct-1 and
monomeric activity but not that of Oct-1 or Oct-2
Oct-4 factors present in P19 extracts form the predomi-
(Scho¨ler et al., 1991; Pesce et al., 1998; Brehm et al.,
nant complexes with the MORE in vitro (Figure 1), sug-
1999; Pesce and Scho¨ler, 2000). MORE and PORE can
gesting that these proteins provide the observed tran-
be also activated by coexpressing Oct-4 and E1A (data
scriptional stimulation in P19 EC cells.
not shown), indicating that E1A recognizes both dimeric
To study the effect of specific Oct proteins, the same
configurations of Oct-4.
reporter plasmids were cotransfected into 293 cells
along with four different expression vectors (Figure 4B).
PORE and MORE Have Different Potential to
First, comparing two dimer binding sites shows that the
Mediate Synergism between Oct-1 and OBF-1
MORE mediates transcriptional activation two to three
An Oct-1 and Oct-2 specific auxiliary activity was dis-
times more efficiently (Oct-2, Oct-4), or at least as good
covered in lymphoid cells, the B cell-specific coactivator
as, the PORE (Oct-6). Second, comparing different Oct
OBF-1 (OCA-B, Bob-1). This coactivator interacts and
factors shows that Oct-4 is the most potent transactiva-
transcriptionally synergizes with octamer site bound
tor in this particular cellular context. In contrast, Oct-1
Oct-1 or Oct-2, but neither with Oct-4 nor Oct-6 (Luo et
was barely able to stimulate either reporter in our trans-
al., 1992; Gstaiger et al., 1995; Luo and Roeder, 1995;
fection experiments (data not shown). Thus, the Oct
Strubin et al., 1995). We investigated whether OBF-1
factors exhibit different strengths in stimulating tran-
can serve as a bridging factor also for Oct-1 and Oct-2
scription although they can all bind to the MORE equally
dimers. As revealed by transient transfection, this co-
well (Figure 2 and Botquin et al., 1998). One possible
activator failed to stimulate MORE-mediated transcrip-
reason is that transcriptional coactivators that act in
tion alone or in conjunction with either Oct-1 or Oct-2
conjunction with Oct-1, Oct-2, and Oct-6 are not ex-
in 293 cells (Figure 5A). In contrast, PORE-mediated
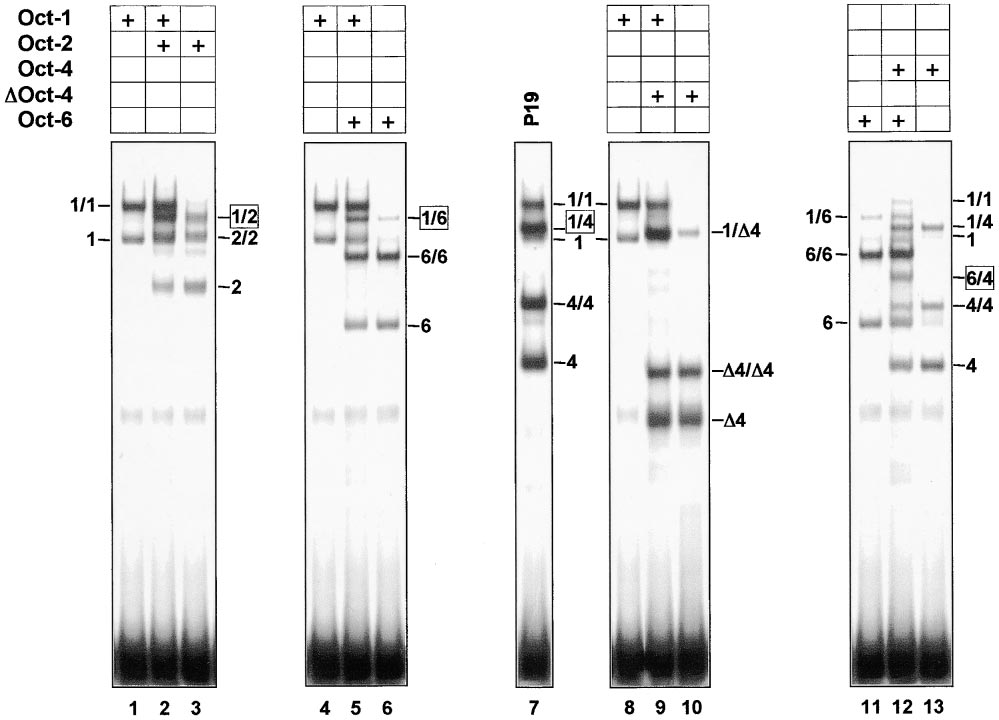
Figure 2. Heterodimerization of Oct Factors on the MORE
293 cells (a human kidney epithelium cell line) were transfected with cytomegalovirus (CMV) promoter-based plasmids directing the expression
of full-length Oct-1 (lanes 1, 4, 8), Oct-2 (lane 3), Oct-6 (lanes 6, 11), Oct-4 (lane 13), and truncated Oct-4 (⌬4, lane 10). The extracts were
mixed in pairs, as indicated above each panel, and subjected to EMSA using the consensus MORE (Figure 1) as a probe. The whole cell
extracts from nontransfected P19 EC cells are also included (lane 7) to show the heterodimer comprised of the endogenous Oct-1 and Oct-4
proteins, seen in Figure 1. The mobility of DNA-protein complexes formed on this gel is marked as per Figure 1.
transcription was significantly stimulated by OBF-1
that the promoter context in OBF-1 recruitment is impor-
tant. On the other hand, the octamer-mutated PORED
cointroduced with either Oct-1 or Oct-2. The level of
activation, ranging from 10- to 35-fold, depended on
mutant (ATTTGAAAgGCAAAT) is indistinguishable from
the ratio of the Oct factor and the OBF-1 protein. The
the natural PORE with regards to OBF-1 recruitment.
observed dependence on the stoichiometry is consis-
Thus, the PORE represent a new class of OBF-1-respon-
tent with OBF-1 acting as a bridging factor binding be-
sive DNA elements that efficiently recruit this coactivator
tween the upstream activator (here Oct-1 or Oct-2) and
through corresponding dimers of Oct-1 (Oct-2) in an
the basal transcription machinery (Scho¨ler et al., 1991).
Lower amounts of exogenous Oct-1 are required to
Further EMSAs revealed a good correlation between
achieve maximum synergy with OBF-1, which could be
the ability of the PORE and PORED (versus MORE and
explained by the fact that 293 cells express endogenous
POREM) to mediate synergism in transcriptional activa-
Oct-1 protein. Furthermore, the Oct-2/OBF-1 pair stimu-
tion in vivo and the ability of these sites to mediate the
lated transcription about 2-fold more than the Oct-1/
interaction between OBF-1 and Oct-1 in vitro (Figure
OBF-1 pair. This may be due to different inherent poten-
5B). Consistent with the reported OBF-1 specificity to
tials of the Oct-1 and Oct-2 transactivation domains
POU domains (Luo and Roeder, 1995; Sauter and Mat-
(Babb et al., 1997).
thias, 1998), OBF-1 neither interacted nor synergized
In order to determine whether OBF-1 prefers mono-
with Oct-4 bound to the PORE (data not shown).
mer or dimer configuration of the POU domain, we in-
The PORE and derivatives thereof were compared to
cluded two PORE-derived sites in the analysis, PORED
the octamer site from the immunoglobulin light chain
and POREM (Figure 5A and Botquin et al., 1998). The
promoter (V, Bergman et al., 1984) that is likely to be
association between OBF-1 and Oct-1 or Oct-2 mono-
a natural target of OBF-1 (Gstaiger et al., 1995; Strubin
mer requires adenosine at the fifth position within the
et al., 1995). Strikingly, the EMSA revealed that the V
classical octamer (ATGCAAAT), or within derivatives
octamer recruits OBF-1 less efficiently than the PORE
thereof (Cepek et al., 1996; Gstaiger et al., 1996). Even
and PORED and only slightly better than the POREM (Fig-
though the POREM represents the canonical octamer
ure 5C). Also unexpected, the mobility of the V complex
sequence with the required adenosine (ATgTGAAATG
is lower compared to those formed on the PORE. It is
CAAAT), this site exhibits virtually no enhancer activity
possible that conditions in our EMSA assay were favor-
(Figure 5A). This result is reminiscent of the failure of
able for the binding of an extra OBF-1 or Oct-1 molecule
OBF-1 to stimulate the histone 2B promoter octamer
to the complex with the V octamer. Another explanation
(Luo and Roeder, 1995). Thus, both sets of data indicate
might be that the PORE-mediated complex induced
OBF-1 Coactivator Synergy Is Mediated by POU Dimer
857
number of genes with these motifs in intronic and pro-
moter regions. For example, the MORE within the ␥-actin
gene first intron (GenBank accession number U20365)
matches the consensus MORE (ATGCATATGCAT). The
MOREs in the Hsp84 gene promoter (ATGCATATGCAa,
number U47056) and in a Bmp4 intron (ATGCATATG
CAg, number D14814) are slightly divergent from the con-
sensus MORE sequence within the POUH docking sub-
sites AT. All three motifs were able to support Oct factor
dimerization in EMSAs (data not shown).
One of the identified homologies, ATGCATATGCAa,
is located within the human Ig VH promoter LR35 (Figure
6A). Strikingly, this MORE lies within a nucleotide stretch
(CTCATGCATATGCAaAT), which differs only in one po-
sition from the sequence referred to for more than a
decade as the heptamer/octamer motif (CTCATGaA
TATGCAaAT; Kemler et al., 1989; LeBowitz et al., 1989;
Poellinger et al., 1989). Further database searches, using
the LR35 MORE plus adjoining promoter sequences as
a query, found the consensus heptamer/octamer motif
itself (e.g., BCL1, Poellinger et al., 1989) and slightly
divergent sequences like B9c (Figure 6A). All these se-
quences are located within Ig VH promoters at the same
distance from the TATA box (Figure 6A). The BCL1- and
B9c-type MOREs occur in numerous human and mouse
VH promoters, whereas the LR35-type MORE appears
to be unique. When these VH MOREs were subsequently
used to search the database, we found MOREs within
the promoter regions of crucial genes like the human
RNA polymerase II gene (ATGAATATGCAg, number
Z54152) or in an intron of the human -globin gene
(ATGAATATGCAa, number U01317.1).
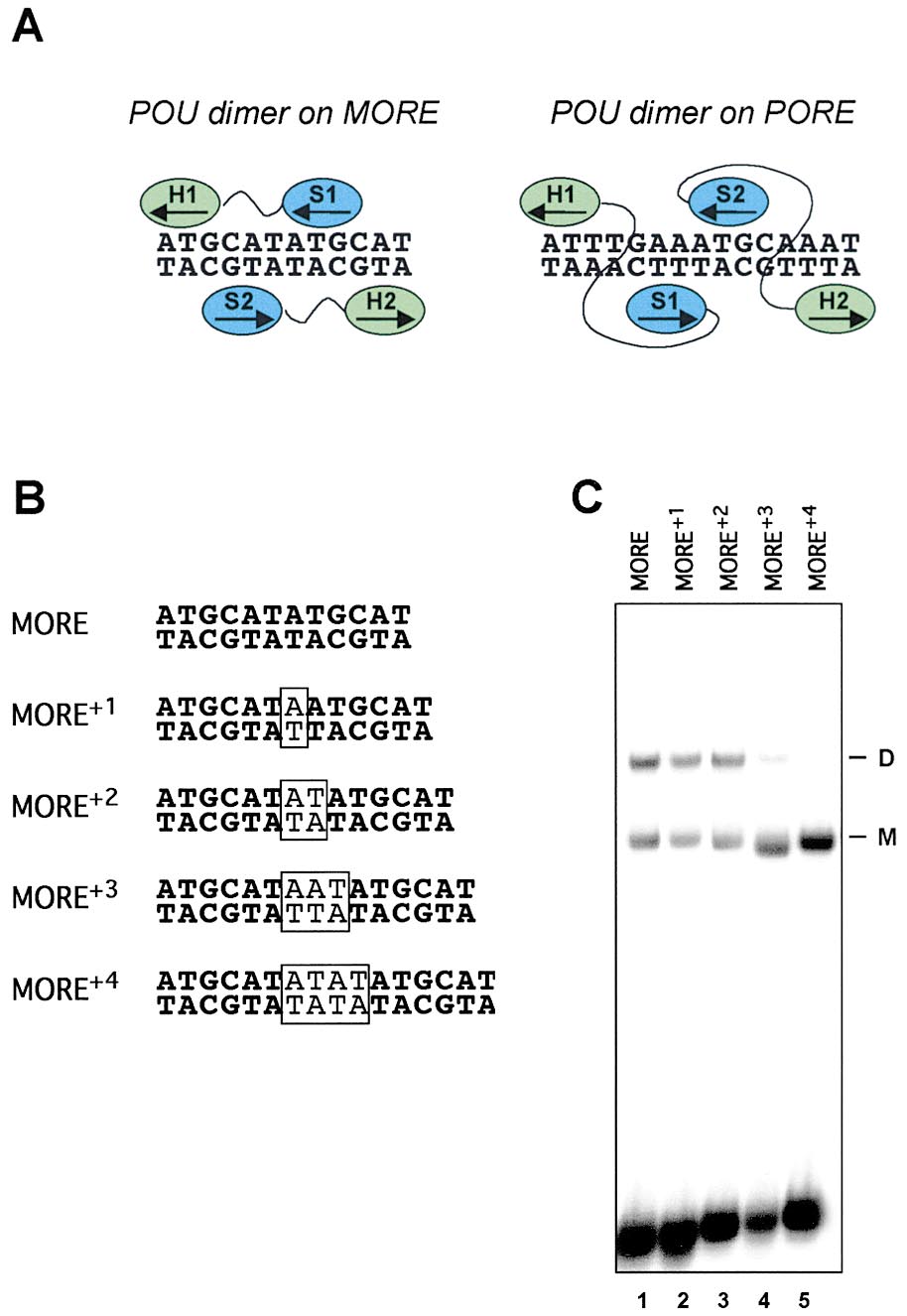
Figure 3. Distinct Configuration of POU Dimers on the MORE and
PORE
We focused our further analysis on the three distinct
Ig VH promoters. One of these promoters, the BCL1, had
(A) Scheme summarizing the overall arrangements of the POU sub-
domains within the Oct-1:MORE crystal structure and in the Oct-
been studied extensively and claimed to be one of the
1:PORE model. The POUS domain (S) and the POUH (H) are indicated
major cis-elements recruiting OBF-1 through octamer
in blue and green, respectively. The POU subdomains belonging to
motif bound Oct-1 or Oct-2 (Luo et al., 1992; Luo and
one polypeptide chain have the same numbering and are connected
Roeder, 1995). The double mutation, Ile159Asp and
by a linker in black. Arrows indicate the direction of the chain from
Asn160Ala, was introduced into the POU domain of
the N to C terminus.
Oct-1. According to the crystallographic data, the resi-
(B) Oligonucleotide probes used to assess the effect of phasing
mutations on the MORE upon dimerization. Inserted base pairs are
dues are located at the C-terminal part of the ␣ helix 3
shown in boxes.
in the POUH domain that forms the MORE-type dimer
(C) EMSA using the MORE plus its phasing mutants (B) as probes,
interface (Figure 7; A. R. et al., unpublished). The indi-
and recombinant Oct-4 as a testing protein. M and D indicate, re-
cated mutation had little effect on the monomer or
spectively, the monomeric and homodimeric Oct-4 complexes with
PORE-type dimer binding, but abolished dimerization on
the consensus MORE (data not shown). It also abolished
dimerization on all three natural MOREs from the VH
conformational changes in the DNA resulting in an in-
promoters (Figure 6B), suggesting the same arrange-
creased mobility evident in the EMSA.
ment of the POU subdomains.
To determine the number of POU molecules within
None of the three VH MOREs was able to efficiently
the complexes supershifted by OBF-1, we performed
mediate an interaction with OBF-1 in EMSAs (Figure 6C).
an EMSA using two differently sized versions of Oct-1
A weak OBF-1 binding can be attributed to the classical
(Figure 5D). OBF-1 supershifted POU-1 and ⌬Oct-1 to
monomeric POU-1 complex (Klemm et al., 1994) formed
different positions, reflecting the different sizes of these
on the overlapping octamer subsite. Indeed, the residual
two Oct-1 species. After mixing all three proteins, a
complex with OBF-1 was eliminated by destroying this
new band between these complexes appeared that was
subsite (mutant BCL1M). In contrast, a significant gain
likely to be a complex of OBF-1 with the POU-1/⌬Oct-1
of OBF-1 binding occurred upon conversion of the
heterodimer (Figure 5D). Thus, this mixing experiment
MORE to the PORE (mutant BCL1P, Figure 6C), consis-
suggests that the OBF-1 complex assembled on the
tent with the previous data on the OPN PORE (Figure 5B).
PORE comprises a POU dimer.
The in vitro data were further correlated with transient
transfection, performed as described in Figure 5A. In
The Heptamer/Octamer Motif Is a MORE Variant
293 cells, the VH MORE-containing promoters (LR35,
A nucleotide database search using MORE and its spac-
B9c, and BCL1) can respond to OBF-1 only weakly, but
ing variant sequences (Figure 3) revealed a significant
even this weak activation is due to the octamer submotif
Figure 4. MORE-Mediated
(A) Comparison of enhancer activities of the
MORE and PORE in transient transfection ex-
periment. P19 EC cells were transfected with
different amounts of luciferase reporter plas-
mids (X axis) containing hexamers arranged
in tandem. The hexamers were copies of the
MORE or PORE plus 10–15 bp flanks from
each side, inserted 37 bp upstream of the
TATA box promoter of the thymidine kinase
(tk) gene. Human -actin LacZ vector (0.1 g)
was included as an internal control of the
transfection efficiency. Y axis: activation of
transcription, expressed as a ratio of lucifer-
ase to -galactosidase activities.
(B) Cotransfection of 293 cells with 0.2 g
of the same reporter plasmids and varying
amounts of CMV-based plasmids (X axis) ex-
pressing Oct-2, Oct-6, or Oct-4. The ⫺37tk-
luc enhancerless vector served as a negative
control in this experiment; the pCMV-lacZ (50
ng) was used for normalization. Y axis: fold
activation, which refers to the background
activity of reporter vectors in cells transfected
with no effector plasmids (the latter taken as
1 for each effector series). The correlation
between protein levels and activation of the
luciferase gene was verified in the EMSA us-
ing extracts of transfected cells (data not
shown).
(cf. BCL1M, Figure 6D). OBF-1 responsiveness was
for the neuronal POU factor Brn-2 (Rhee et al., 1998).
achieved by converting the BCL1 MORE to the PORE
The POUS domains of these POU proteins exhibit distinct
within the same promoter context (BCL1P). Very similar
sequence requirements. Moreover, the POUS domain of
results were obtained with the LR35M and LR35P deriva-
a given POU protein can alter its interaction with DNA
tives of the corresponding VH promoter (data not shown).
in the MORE dimeric configuration, allowing recognition
Thus, the outcome of the transfection experiment (Fig-
of divergent sequences. For example, the POUS domain
ure 6D) is in agreement with the obtained in vitro results
of Oct-1 can bind the ATGC (LR35 MORE) and ATTC
(Figure 6C). Our data show that the coactivator OBF-1
(BCL1 MORE) subsites equally well (Figure 6B). The
cannot be recruited efficiently to the VH promoter bound
specificity of DNA binding by the POUH subdomains
Oct-1, which is in contrast to a view commonly accepted
in the MORE dimeric configuration is relaxed as well:
so far (Luo et al., 1992; Luo and Roeder, 1995).
besides the AT subsites, Oct-1 POUH can recognize the
AA (VH and Hsp84 MOREs) or AG (Bmp4 MORE). Finally,
the MORE-type configuration tolerates variable spacing
between POUS docking subsites (Figure 3 and Rhee et
al., 1998). Taken together, these data suggest that there
MORE-Mediated Dimerization Is Universal
is a wide range of possible in vivo complexes where
for POU Domains
divergent POU domains assemble on divergent DNA
The consensus MORE used for crystallographic studies
sequences in a MORE-like fashion.
appears to be an affiliate of a broad class of similar DNA
elements. This class includes the prolactin/PitD site (Ja-
Heterodimerization on the MORE
cobson et al., 1997) and, likely, the motif ATG(C/A)AT
A remarkable feature of the MORE resides in its ability
(A/T)0–2ATTCAT that is the optimal dimerization substrate
to enable homo- and heterodimerization of a variety of
OBF-1 Coactivator Synergy Is Mediated by POU Dimer
859
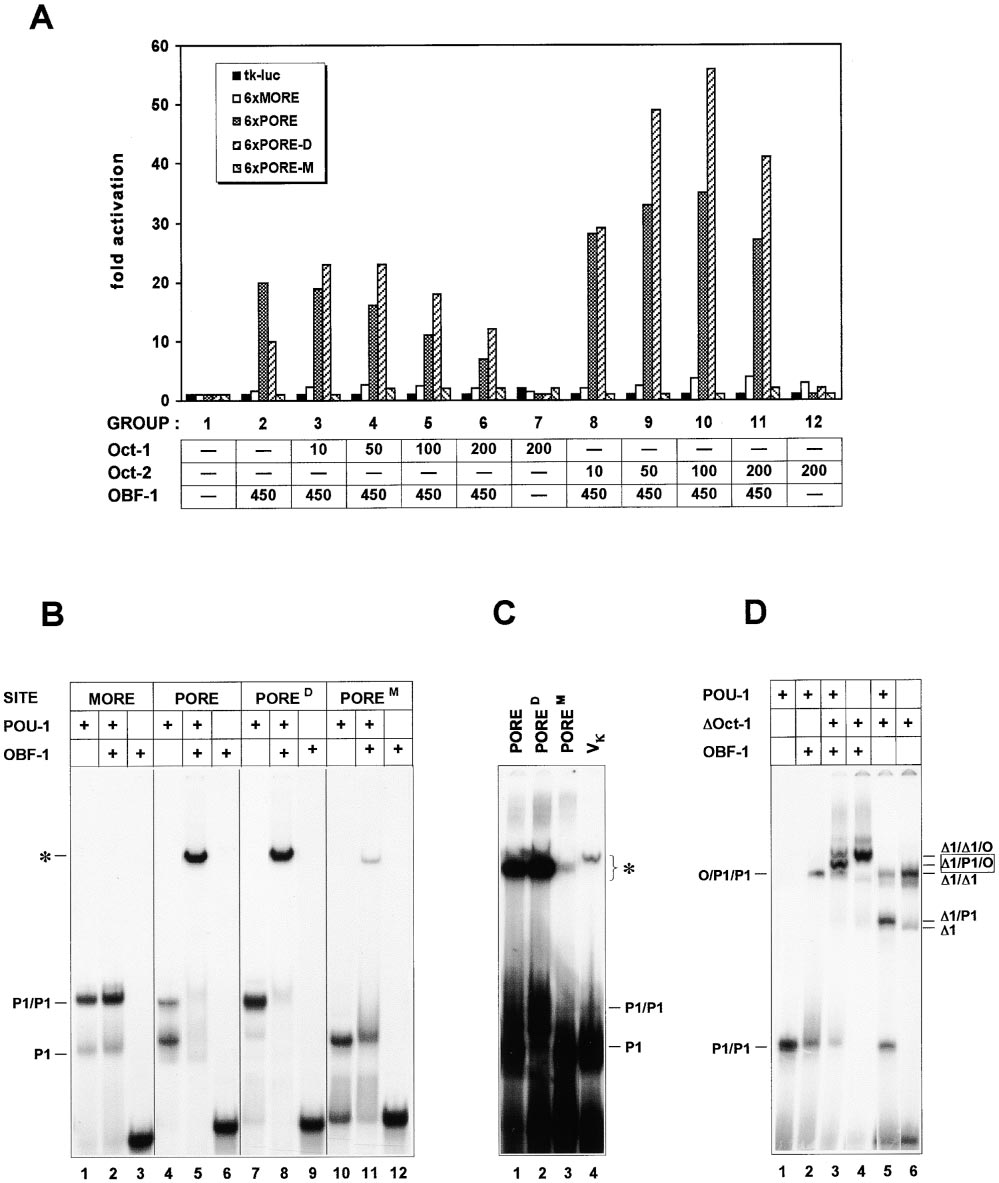
Figure 5. Selective Recruitment of the Coactivator OBF-1 to the POU Dimer Formed on the PORE
(A) Transient transfection of 293 cells. The luciferase reporter plasmids 6xMORE and 6xPORE were described in Figure 4. The PORED
(ATTTGAAAgGCAAAT) and POREM (ATgTGAAATGCAAAT) are mutants of the PORE that were purposely designed to selectively bind Oct
factor dimers and monomers, respectively (Botquin et al., 1998). Cells were cotransfected with CMV-based Oct-1, Oct-2, and OBF-1 effector
plasmids (nanograms, X axis). Fold activation (Y axis) refers to the luciferase activity in cells transfected with no effector plasmids (group 1).
The pCMV-lacZ vector (50 ng) was used for standardization.
(B) The bacterially produced POU-1 (see Figure 1B) and OBF-1 proteins were tested in EMSA using the 32P-labeled MORE, PORE, and mutated
versions thereof (PORED and POREM) as probes. P1 and P1/P1 refer to the POU-1 monomer and dimer, respectively, and the POU-1:DNA
complex that is supershifted by OBF-1 is marked by asterisk.
(C) PORE and its mutants were compared in EMSA to the octamer site of the immunoglobulin kappa light chain promoter (V). The POU-
1:DNA complexes that are supershifted by OBF-1 are denoted by asterisk without specifying the number of POU-1 and OBF-1 molecules
therein. The abbreviations used are the same as in (B).
(D) The OBF-1/Oct-1:PORE complex contains two POU domain molecules. ⌬Oct-1 protein (⌬1), described in the legend of Figure 3, was
introduced in the analysis in addition to POU-1 (P1) and OBF-1 (O). The proteins were mixed in different combinations, as indicated above
the panels, with the labeled PORED probe and subjected to EMSA. Notice the appearance of an intermediate species (⌬1/P1/O, lane 3), which
likely represents the POU-1/⌬Oct-1 heterodimer (⌬1/P1, lane 5) that is supershifted by OBF-1.
Oct factors (Figure 2). This is surprising considering that
Oct-6 (positions 159 and 160). Nevertheless, a computer
some amino acids making up the MORE-type dimeriza-
modeling of corresponding subdomains into the coordi-
tion interface are quite variable among the Oct factors
nates of the Oct-1:MORE crystal demonstrates that
(for alignment of the POU domains see Herr and Cleary,
these amino acids can fit into the conserved hydropho-
1995). Such are, for example, the two last amino acids
bic pocket of the interacting POUS domains (Figure 7B
of the ␣ helix 3 in the POUH domains of Oct-1, Oct-4, and
and A. R. et al., unpublished).
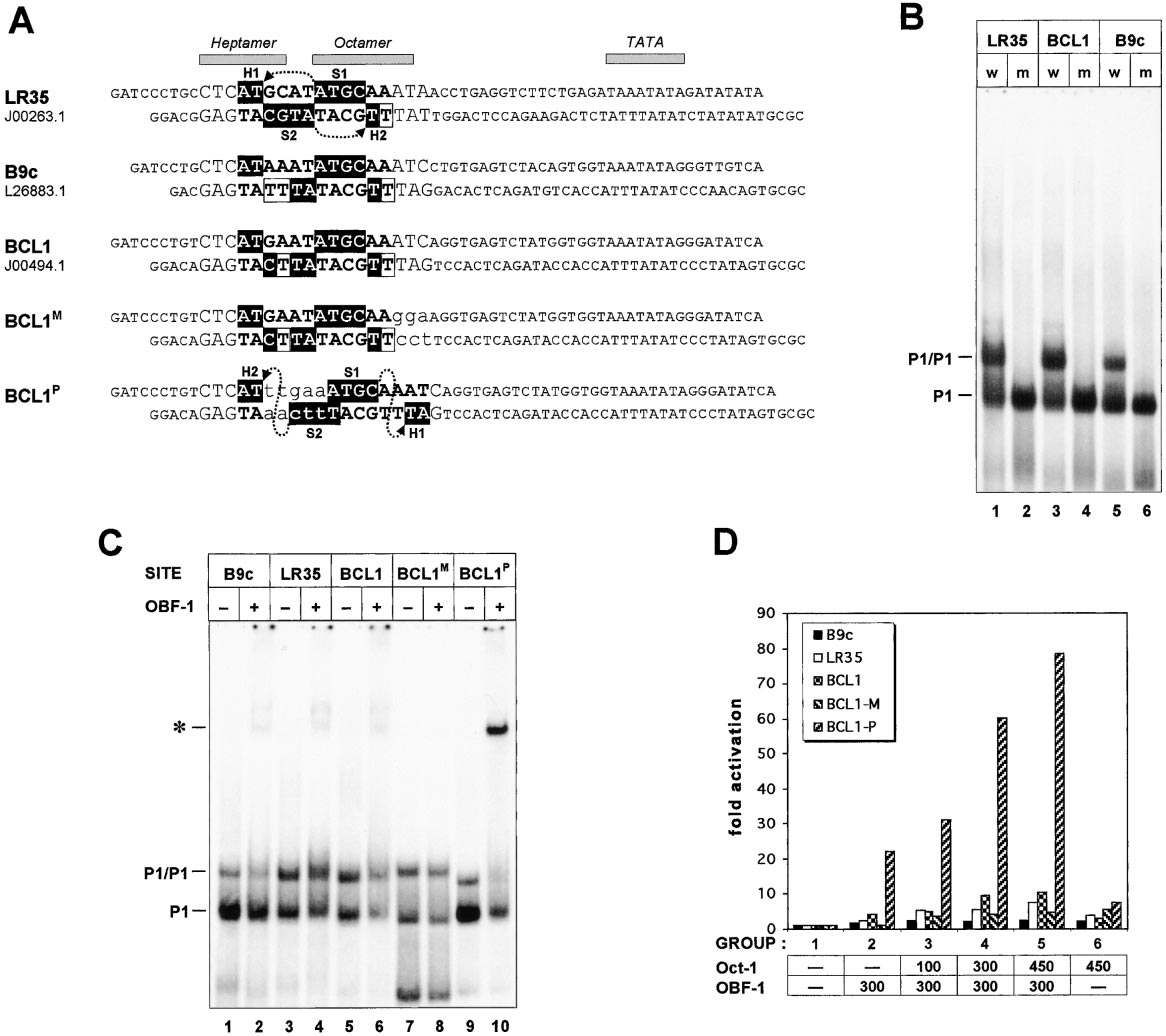
Figure 6. OBF-1 Cannot Be Recruited to the Oct-1 Dimers Bound to Natural MOREs from the Immunoglobulin Heavy Chain Promoters (VH)
(A) Three representative VH promoters containing slightly different MOREs (upper three sequences). Numbers under the names specify GenBank
entries these sequences were retrieved from. LR35 is unique, whereas the BCL1 is the most abundant type of the MOREs occurring in VH
promoters family. In the MOREs (shown in bold) and PORE (last lane), the docking sites for the POU subdomains are in filled boxes; open
boxes in the VH MOREs designate positions, respectively, matching to and divergent from the consensus MORE (ATGCATATGCAT). The
mutations were introduced in the BCL1 promoter fragment (two bottom sequences): in the BCL1M (MORE) the octamer part was destroyed (lower
case) without affecting the MORE itself, and in the BCL1P (PORE) mutant, the MORE was converted to the PORE. Note the distinct arrangements
of the POU subdomains on the MOREs and PORE (see also Figure 3A). The nucleotide stretches previously referred to as heptamer and
octamer motifs, and the TATA boxes are denoted on the top; the BamHI and MluI half-sites at the ends were introduced to facilitate cloning.
(B) EMSA analysis of bacterially produced wild-type (w) and mutated (m: Ile159Asp, Asn160Ala) forms of the Oct-1 POU domain (P1) using
32P-labeled VH promoter fragments (A) as probes.
(C) EMSA with the wild-type Oct-1 POU domain and OBF-1 proteins. Oligonucleotide probes are described in (A); the asterisk points to the
OBF-1/POU-1:DNA complex.
(D) Transient transfection of the 293 cells. By cloning the VH promoter fragments (A) upstream of the transcription start of the luc gene, the
reporter plasmid series was created. The assay conditions, effector plasmids, and abbreviations are as in Figure 5A.
MORE Dimerization Prevents OBF-1 Recruitment
was the first regulatory DNA element reported to medi-
to VH Promoters: Correlation with OBF-1
ate interaction and synergism between Oct-1 (Oct-2)
Deficiency in Mice
and OBF-1 (Luo et al., 1992). This apparent discrepancy
The consensus MORE used for X-ray crystallography
can be explained considering the deletion of the hep-
(ATGCATATGCAT, A. R. et al., unpublished) and the
tamer subpart and consequently, the ablation of MORE-
MOREs from the Ig VH promoters (AT[G/a][C/a]ATATG
mediated dimerization of Oct-1 (Oct-2) in the DNA con-
CAa, Figure 6A) bind the Oct-1 dimer in the same config-
structs used in those studies (Luo et al., 1992; Luo and
uration (Figure 6B) hampering the recruitment of the
Roeder, 1995). Although this deletion allowed isolation
coactivator OBF-1 (Figures 6C and 6D). The VH MOREs
and characterization of OBF-1, it also led to the con-
are included in the well-known heptamer/octamer motifs
clusion that this protein activated Ig gene transcription
(Kemler et al., 1989; LeBowitz et al., 1989; Poellinger et
via the octamer motif within the VH promoter. To illus-
al., 1989), and one of them (from the BCL1 VH promoter)
trate this situation, we mutated the BCL1 MORE
OBF-1 Coactivator Synergy Is Mediated by POU Dimer
861
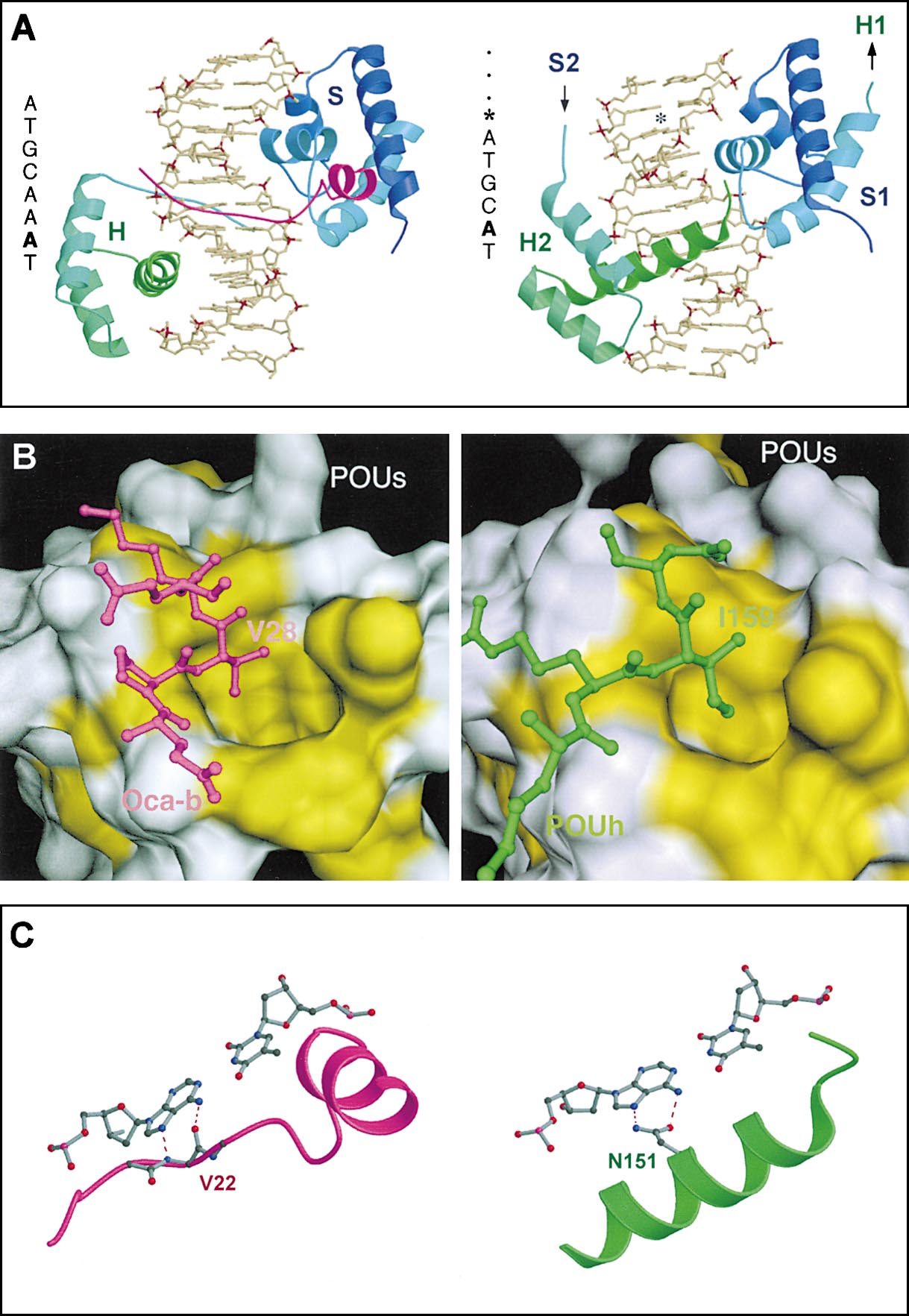
Figure 7. The Recognition Helix of the POUH
in the Oct-1:MORE Crystal Structure and the
OBF-1 Peptide in the Ternary Oct-1/OBF-
1:Octamer Motif Complex Occupy the Same
Position in the Major Groove of the DNA
(A) Overall structure of the Oct-1:MORE and
the Oct-1/OBF-1:octamer complexes. Left
panel: the crystal structure of the ternary
complex between the Oct-1 POU domain, the
OBF-1 peptide, and the octamer binding site.
The coordinates of Chasman et al. (1999)
were used for this figure. In this structure, the
Oct-1 POU domain is bound as a monomer;
the linker (not shown) connects POUS (S) and
POUH (H) subdomains. Right panel: The crys-
tal structure of the Oct-1:MORE dimer com-
plex. Only one half of the complex bound to
one half-site of the palindromic MORE is
shown. The POUS (S1) and POUH (H2) subdo-
mains in this half complex originate from two
different polypeptide chains. The complete
dimer image is generated by a 2-fold rotation
along an axis positioned between the two
half-sites of the palindromic MORE that is
perpendicular to the plane of the figure (ATG
CAT*ATGCAT, indicated by asterisk). Color
code: POUS domain, blue; POUH domain,
green; OBF-1 fragment, magenta. The bright-
ness of the colors is ramped from the N termi-
nus to the C terminus of each domain. The
DNA motifs of the two complexes are given
to the left of each panel. The base pair in the
5th position is highlighted in bold (see C). In
the ternary Oct-1/OBF-1:octamer complex
the OBF-1 peptide makes extensive protein–
protein interactions with the POUS domain
and contacts the POUH via the major groove
in the DNA (left). There is a change in spacing,
by two base-pairs, in the DNA binding of the
POUH domain in the Oct-1:MORE dimeric
structure compared to that of the Oct-1/OBF-
1:octamer complex.
(B) The intermolecular hydrophobic interac-
tions in the Oct-1/OBF-1:octamer and in the
Oct-1:MORE complex are similar in nature.
The protein–protein interface in the Oct-1/
OBF-1:octamer motif (left panel) and in the
Oct-1:MORE complex (right panel) are com-
pared. The interface of POUS is shown as a surface presentation. The hydrophobic amino acids that are involved in the interface are colored
in yellow. Segments of OBF-1 (magenta) and POUH (green) are shown in ball-and-stick presentation. Both Val28 (OBF-1) and Ile159 (POUH)
bind to the same hydrophobic pocket of the POUS domain.
(C) The A:T base pair in the 5th position plays a key role in the protein–DNA interface in the ternary Oct-1/OBF-1:octamer complex (left) and
in the Oct-1:MORE complex (right). The backbone of the OBF-1 peptide (Val22) provides a pair of hydrogen bonding interactions with the
adenine base in the 5th position of the octamer motif (left). The same base in the MORE binding site is hydrogen bonded with the side chain
of Asn151 from the recognition helix of the POUH domain (right).
(cagggTATGCAAAT) with the purpose to eliminate the
6; see next section for the underlying structural aspects).
dimer assembly without disturbing the monomer binding
However, it is possible that, under certain physiological
to the octamer site. The indicated mutation created a
conditions, dimerization is prevented, e.g., by phosphor-
strong (similar to the V octamer) OBF-1 responsive
ylation, and an Oct-1 or Oct-2 monomer binding to the
enhancer, although transcriptional activity is weaker
high-affinity octamer sequence becomes accessible to
than for the PORE (data not shown).
The phenotype of OBF-1-deficient mice eventually
Deficiency of OBF-1 does have an impact on Ig gene
challenged the idea about direct recruitment of OBF-1
transcription, but only subsequent to immunoglobulin
to the VH promoters. It was shown that the transcription
class switch in antigen-responding B cells. Secondary
of Ig genes in OBF-1-deficient mice was largely unaf-
heavy chain Ig isotypes are expressed at severely re-
fected (Kim et al., 1996; Schubart et al., 1996). Our study
duced levels in OBF-1⫺/⫺mice (Kim et al., 1996; Schubart
provides a rationale for this: the dimerization of Oct-1
et al., 1996). The immunoglobulin class switch, charac-
on the VH MOREs does not allow the recruitment of
terized by the recombination of the VDJ to C region,
OBF-1 to the corresponding promoters (Figures 5 and
brings the 3⬘-IgH enhancer into proximity to the VH pro-
moters. Remarkably, this enhancer contains no MORE-
like sequences but the consensus octamer motif. As
A hallmark of the POU domain family transcription fac-
opposed to the VH promoters, the latter appears to be
tors is their flexibility in DNA recognition (reviewed by
a bona fide genomic target for OBF-1 (Stevens et al.,
Herr and Cleary, 1995). In this study, we show that the
flexibility in POU factor functioning can also be extended
to dimerization. We demonstrate the binding of Oct fac-
The Structural Basis for Differential
tor family members as homo- and heterodimers to the
Recruitment of OBF-1
two high-affinity regulatory elements, the PORE and the
The crystal structure of a POU complex in the PORE
MORE. The structural difference between PORE- and
dimer configuration without OBF-1 became available
MORE-mediated dimerization leads to the differential
just recently (A. R. et al., unpublished). Preliminary crys-
recruitment of transcriptional coactivators. OBF-1, for
tallographic data revealed an arrangement of the POU
example, binds and synergizes in transcriptional activa-
subdomains very similar to that predicted by computer
tion with the PORE configuration of the Oct-1 dimer, but
modeling (Figure 3 and Botquin et al., 1998), providing
fails to bind to the MORE-mediated Oct-1 dimer. Thus,
an idea of the structural basis of this coactivator interac-
our data demonstrate the mechanism by which distinct
tion in the PORE dimer. Since the PORE structure is
POU dimer configurations can recruit specific transcrip-
based on the monomer configuration in the Oct-1:oc-
tional coactivators with different effects on gene tran-
tamer crystal structure (Klemm et al., 1994), we assume
scription. In addition, we outline the structural parame-
that the observed binding surface of OBF-1 in the mono-
ters leading to this selectivity in coactivator recruitment.
mer (Chasman et al., 1999) is the same in the PORE
The Ig VH promoter fragments, containing the MOREs
(Figure 6A), have been shown to be fairly active in B
The ternary monomer complex shows the way the
cells or B cell extracts (Kemler et al., 1989; LeBowitz et
OBF-1 fragment binds to the N-terminal part of helix 1
al., 1989; Poellinger et al., 1989). Since OBF-1 fails to
(residues 6–10) and a segment between helices 3 and
activate these promoters (Figure 6D), it is tempting to
4 (residues 49–60) of the POUS domain (Figure 7A, left).
speculate that a yet unknown class of transcription co-
Our new structure of the Oct-1:MORE dimer complex
regulators exists. This novel class should have an oppo-
(Figure 7A, right and A. R. et al., unpublished) provides
site specificity for dimer assembly specifically binding
a rationale for why binding of OBF-1 is inhibited in this
to the MORE-type configuration of the POU domain.
dimer configuration. The direct comparison of the Oct-1/
OBF-1:octamer complex (Figure 7A, left) and the Oct-1:
MORE dimer (Figure 7A, right) reveals that the binding
site for OBF-1 is identical to the protein–protein POUS/
PORE-, PORED-, and POREM-containing oligonucleotides were de-
POUH interface site in the MORE dimer, in which the
scribed by Botquin et al. (1998), named there O, O⫺1, and O⫺3,
same residues of POUS (helix 1 and the loop between
respectively. The consensus MORE, its spacing derivatives, and
helices 3 and 4) interact with the C terminus of POUH.
PitD motif (indicated with X) were placed into the PORE-flanking
The most important contact within this interface is a
regions derived from the OPN intron (upper strand: 5⬘-CTGAAAGT
key–lock type interaction: the side chain of Ile159 of
POUH fits into a hydrophobic cavity of POUS (Figure
placement (underlined) in a flanking region was required to eliminate
7B, right). The equivalent interaction is observed in the
an occasionally created binding site for an unknown protein from
Oct-1/OBF-1:octamer complex where Val28 of OBF-1
the 293 cell extracts. The sequence of the V octamer-containing
fits into the very same pocket of the POUs domain of
oligonucleotide is as follows: 5⬘-CTGACTCCTGCCTTCAGGGTATG
Oct-1 (Figure 7B, left).
The analogy can be further extended to specific DNA
base binding (Figure 7C). The amido group of the Asn151
The CTGA and TCAG 5⬘-protruding sequences are of nongenomic
origin.
side chain of POUH makes two specific hydrogen bonds
to the A:T base pair in position 5 of the MORE (Figure
7C, right). This hydrogen bond interaction is regarded
The POU domain of Oct-1 (POU-1) was amplified from its cDNA by
as a signature for DNA binding of homeo domains
(Brehm et al., 1998). In the Oct-1/OBF-1:octamer com-
plex, the same base is hydrogen-bonded by the amino
GCG-3⬘ oligonucleotides. The amplified fragment was first cleaved
group and by the carbonyl group of the main chain of
with NcoI and NotI, then cloned into pET9d-NHis6 vector (which
is a modified version of pET-9d(⫹) from Novagen). The resulted
Val22 of OBF-1 (Figure 7C, left). From this structural
construct was used for a site-directed mutagenesis to create a
comparison, we conclude that OBF-1 and the POUH
vector expressing the POU-1 mutant Ile159Asp, Asn160Ala. The full-
domain compete for binding to the same site of the
length Oct-4 cDNA was PCR-amplified with oligonucleotides:
POUS domain where in the MORE dimer, the OBF-1
binding site is blocked by POUH but accessible in the
CAGAGGGAACCTCCTCTGAG-3⬘ and ligated into the pCR2.1 vector
predicted PORE dimer. The specificity of competitive
(TA cloning kit, Invitrogen). The insert was cleaved out by NcoI and
binding of OBF-1 and POU
NotI and ligated into pET9d-NHis6. The primers 5⬘-GGGAGGACGT
H is further enhanced by the
capability of the two competing domains, POUH and
GCCGCTAAAAGCCCTCCACGGAGAGG-3⬘ were used to amplify the
OBF-1, to specifically interact with the binding motif of
full-length OBF-1. The generated fragment was cleaved with BspH1/
the respective DNA. The data also indicate that the
NotI and ligated into NcoI/NotI-linearized pET24-TEV-His6 plasmid,
POUS/POUH binding affinity of the examined MORE di-
which was a derivative from pET-24d (Novagen). The truncated
mer complexes is superior compared to the affinity of
Oct-1 (⌬Oct-1, amino acids 183–508) was generated from cDNA
OBF-1 Coactivator Synergy Is Mediated by POU Dimer
863
factor Oct-4: viral oncoproteins share the ability to mimic a stem
otides. The fragment was cleaved with NcoI/SacI and cloned into
cell-specific activity. Mol. Cell. Biol. 19, 2635–2643.
pET24d-TEV-His6 at corresponding sites.
Cepek, K.L., Chasman, D.I., and Sharp, P.A. (1996). Sequence-spe-
The cytomegalovirus (CMV) promoter-driven eukaryotic expres-
cific DNA binding of the B-cell-specific coactivator OCA-B. Genes
sion plasmids were obtained from other investigators (pCG-Oct1, W.
Dev. 10, 2079–2088.
Herr; pEV-OBF1, P. Matthias), and pCMV-Oct2, pCMV-Oct4, pCMV-
Chasman, D., Cepek, K., Sharp, P.A., and Pabo, C.O. (1999). Crystal
16NOct4 (here referred to as ⌬Oct-4), and pCMV-Oct6 were de-
structure of an OCA-B peptide bound to an Oct-1 POU domain/
scribed previously (Scho¨ler et al., 1990; Brehm et al., 1997). The
octamer DNA complex: specific recognition of a protein-DNA inter-
MORE-containing oligonucleotides (see above) were multimerized
face. Genes Dev. 13, 2650–2657.
and cloned into the ⫺37tk-luc enhancerless vector in the same way
Gstaiger, M., Knoepfel, L., Georgiev, O., Schaffner, W., and Hovens,
as the PORE reporter series was generated (Botquin et al., 1998).
C.M. (1995). A B-cell coactivator of octamer-binding transcription
The B9c, LR35, and BCL1 reporter series were created by replacing
factors. Nature 373, 360–362.
the BamHI/MluI promoter fragment of the ⫺37tk-luc with the VH
oligonucleotides shown in Figure 6A.
Gstaiger, M., Georgiev, O., van Leeuwen, H., van der Vliet, P., and
Schaffner, W. (1996). The B cell coactivator Bob1 shows DNA se-
quence-dependent complex formation with Oct-1/Oct-2 factors,
leading to differential promoter activation. EMBO J. 15, 2781–2790.
All recombinant proteins were expressed in BL21(DE3)pLysS E. coli
strain (Novagen) and subsequently purified on the Ni-NTA agarose
Herr, W., and Cleary, M.A. (1995). The POU domain: versatility in
columns (Qiagen). The eluted protein solutions were used for the
transcriptional regulation by a flexible two-in-one DNA-binding do-
EMSA that was performed as previously described (Sylvester and
main. Genes Dev. 9, 1679–1693.
Scho¨ler, 1994). Oct-1 antibody was purchased from Santa Cruz
Jacobson, E.M., Li, P., Leon-del-Rio, A., Rosenfeld, M.G., and Ag-
Biotechnology and the generation of Oct-4 polyclonal antibody was
garwal, A.K. (1997). Structure of Pit-1 POU domain bound to DNA
described elsewhere (Palmieri et al., 1994).
as a dimer: unexpected arrangement and flexibility. Genes Dev. 11,
Transient transfection experiments were performed in 24 well tis-
sue culture plates using FuGene6 transfection reagent (Roche). The
Kemler, I., Schreiber, E., Muller, M.M., Matthias, P., and Schaffner,
total amount of DNA per well was equalized to 1 g with a carrier
W. (1989). Octamer transcription factors bind to two different se-
plasmid. After 36–48 hr, cells were washed with PBS, and were
quence motifs of the immunoglobulin heavy chain promoter. EMBO
lysed directly in the wells in 250 mM Tris-HCl (pH 7.8), 1 mM DTT
J. 8, 2001–2008.
through three cycles of freezing in liquid nitrogen and quick thawing
Kim, U., Qin, X.-F., Gong, S., Stevens, S., Luo, Y., Nussenzweig, M.,
in a 37⬚C water bath. Approximately 1/20 of the amount of the crude
and Roeder, R.G. (1996). The B-cell-specific transcription coactiva-
lysate was used to measure the luciferase and -galactosidase ac-
tor OCA-B/OBF-1/Bob-1 is essential for normal production of immu-
tivities in standard assays.
noglobulin isotypes. Nature 383, 542–547.
Klemm, J.D., Rould, M.A., Aurora, R., Herr, W., and Pabo, C.O. (1994).
Crystal structure of the Oct-1 POU domain bound to an octamer
site: DNA recognition with tethered DNA-binding modules. Cell 77,
We are grateful to Patrick Matthias for the pEV-OBF1 vector and
nice and helpful discussions, Winship Herr for pCG-Oct1, and An-
LeBowitz, J.H., Clerc, R.G., Brenowitz, M., and Sharp, P.A. (1989).
dreas Hecht for the ⫺37tk-luc plasmid. We thank Vale´rie Botquin
The Oct-2 protein binds cooperatively to adjacent octamer sites.
for advice, Karin Hu¨bner and Selma Dejgaard for preparation of the
Genes Dev. 3, 1625–1638.
Oct-4 antibody, Stefan Schlatt and Areti Malapetsa for editing the
manuscript, Nathalie Daigle, Konstantinos Anastassiadis, and other
Luo, Y., and Roeder, R.G. (1995). Cloning, functional characteriza-
laboratory members for helpful discussion and support. This work
tion, and mechanism of action of the B-cell-specific transcriptional
was initiated at the EMBL in Heidelberg, and finished at the EMBL
coactivator OCA-B. Mol. Cell. Biol. 15, 4115–4124.
in Hamburg and at the New Bolton Center at the University of Penn-
Luo, Y., Fujii, H., Gerster, T., and Roeder, R.G. (1992). A novel B cell-
sylvania. The work in Heidelberg and Hamburg was supported by
derived coactivator potentiates the activation of immunoglobulin
the EMBL. The work at the University of Pennsylvania was supported
promoters by octamer-binding transcription factors. Cell 71,
by the Jones fund and the Commonwealth of Pennsylvania grants
to H. R. S.
Mangelsdorf, D.J., and Evans, R.M. (1995). The RXR heterodimers
and orphan receptors. Cell 83, 841–850.
Received June 13, 2000; revised November 7, 2000.
Palmieri, S.L., Peter, W., Hess, H., and Scho¨ler, H.R. (1994). Oct-4
transcription factor is differentially expressed in the mouse embryo
during establishment of the first two extraembryonic cell lineages
involved in implantation. Dev. Biol. 166, 259–267.
Babb, R., Cleary, M.A., and Herr, W. (1997). OCA-B is a functional
Pesce, M., and Scho¨ler, H.R. (2000). Oct-4: control of totipotency
analog of VP16 but targets a separate surface of the Oct-1 POU
and germline determination. Mol. Reprod. Dev. 55, 452–457.
domain. Mol. Cell. Biol. 17, 7295–7305.
Pesce, M., Gross, M., and Scho¨ler, H.R. (1998). In line with our
Bergman, Y., Rice, D., Grosschedl, R., and Baltimore, D. (1984). Two
ancestors: Oct-4 and the mammalian germ. Bioessays 20, 722–732.
regulatory elements for immunoglobulin light chain gene expres-
Poellinger, L., Yoza, B.K., and Roeder, R.G. (1989). Functional co-
sion. Proc. Natl. Acad. Sci. USA 81, 7041–7045.
operativity between protein molecules bound at two distinct se-
Botquin, V., Hess, H., Fuhrmann, G., Anastassiadis, C., Gross, M.K.,
quence elements of the immunoglobulin heavy-chain promoter. Na-
Vriend, G., and Scho¨ler, H.R. (1998). New POU dimer configuration
ture 337, 573–576.
mediates antagonistic control of an osteopontin preimplantation
Rhee, J.M., Gruber, C.A., Brodie, T.B., Trieu, M., and Turner, E.E.
enhancer by Oct-4 and Sox-2. Genes Dev. 12, 2073–2090.
(1998). Highly cooperative homodimerization is a conserved prop-
Brehm, A., Ohbo, K., and Scho¨ler, H.R. (1997). The carboxy-terminal
erty of neural POU proteins. J. Biol. Chem. 273, 34196–34205.
domain of Oct-4 acquires cell specificity through the POU domain.
Ryan, A.K., and Rosenfeld, M.G. (1997). POU domain family values:
Mol. Cell. Biol. 17, 154–162.
flexibility, partnerships, and developmental codes. Genes Dev. 11,
Brehm, A., Ovitt, C.E., and Scho¨ler, H.R. (1998). Oct-4: more than
just a POUerful marker of the mammalian germline? APMIS 106,
Sauter, P., and Matthias, P. (1998). Coactivator OBF-1 makes selec-
tive contacts with both the POU-specific domain and the POU ho-
Brehm, A., Ohbo, K., Zwerschke, W., Botquin, V., Jansen-Du¨rr, P.,
meodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol.
and Scho¨ler, H.R. (1999). Synergism with germ line transcription
Scho¨ler, H.R. (1991). Octamania: the POU factors in murine develop-
ment. Trends Genet. 7, 323–329.
Scho¨ler, H.R., Hatzopoulos, A.K., Balling, R., Suzuki, N., and Gruss,
P. (1989). A family of octamer-specific proteins present during
mouse embryogenesis: evidence for germline-specific expression
of an Oct factor. EMBO J. 8, 2543–2550.
Scho¨ler, H.R., Dressler, G.R., Balling, R., Rohdewohld, H., and Gruss,
P. (1990). Oct-4: a germline-specific transcription factor mapping
to the mouse t-complex. EMBO J. 9, 2185–2195.
Scho¨ler, H.R., Ciesiolka, T., and Gruss, P. (1991). A nexus between
Oct-4 and E1A: implications for gene regulation in embryonic stem
cells. Cell 66, 291–304.
Schubart, D.B., Rolink, A., Kosco-Vilbois, M.H., Botteri, F., and Mat-
thias, P. (1996). B-cell-specific coactivator OBF-1/OCA-B/Bob-1 re-
quired for immune response and germinal centre formation. Nature
383, 538–541.
Stevens, S., Ong, J., Kim, U., Eckhardt, L.A., and Roeder, R.G. (2000).
Role of OCA-B in 3⬘-IgH enhancer function. J. Immunol. 164, 5306–
5312.
Strubin, M., Newell, J.W., and Matthias, P. (1995). OBF-1, a novel
B cell-specific coactivator that stimulates immunoglobulin promoter
activity through association with octamer-binding proteins. Cell 80,
497–506.
Sylvester, I., and Scho¨ler, H.R. (1994). Regulation of the Oct-4 gene
by nuclear receptors. Nucleic Acids Res. 22, 901–911.
Verrijzer, C.P., Alkema, M.J., van Weperen, W.W., van Leeuwen,
H.C., Strating, M.J., and van der Vliet, P.C. (1992). The DNA binding
specificity of the bipartite POU domain and its subdomains. EMBO
J. 11, 4993–5003.
Verrijzer, C.P., and van der Vliet, P.C. (1993). POU domain transcrip-
tion factors. Biochim. Biophys. Acta 1173, 1–21.
Voss, J.W., Wilson, L., and Rosenfeld, M.G. (1991). POU domain
proteins Pit-1 and Oct-1 interact to form a heteromeric complex
and can cooperate to induce expression of the prolactin promoter.
Genes Dev. 5, 1309–1320.
Source: http://www.remilab.hu/cikkek/RA/cell.pdf
June - August 2004 EUROPEAN COMMUNITY LAW NEWS DIRECTIVE ON TAKEOVER BIDS offeree company; (ii) sufficient time and European Parliament and Council Directive sufficient information provided to the 2004/25/EC of 21 April 2004 on takeover bids addressees of the bid; (iii) the board of the offeree company acting in the interests of Directive 2004/25/EC of 21 April 2004 on
HORMONES 2004, 3(4):244-251 A Paradox: The Roles of Inositolphosphoglycans in mediating insulinsensitivity and Hyperandrogenism in the Polycystic Ovary Syndrome Kai I. Cheang1, Paulina Essah2, John E. Nestler2,3 1Ph.D., Department of Pharmacy, 2M.D., Department of Internal Medicine, 3M.D., Department ofObstetrics and Gynecology, Medical College of Virginia Campus, Virginia CommonwealthUniversity, Richmond, Virginia













