Scienceindoors.com

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
Drug design strategies using non nucleoside reverse transcriptase inhibitors
(NNRTI): current challenges and future perspectives
Ravi Sharma1, Gaurav Kumar2, Preeti Saini3, Hardeep Singh Tuli4*, Shivani Sood4
1Department of Biotechnology, Mata Gujri Collage, Fatehgarh Sahib, India
2Department of Biochemistry, Kurukshetra University, Kurukshetra-136119, India
3Department of Microbiology, PAU, Ludhiana-141004, India
4Department of Biotechnology, Maharishi Markandeshwar University, Mullana-Ambala-133207
*Correspondence at [email protected]
Received: September 13, 2014; Accepted: November 06, 2014
Abstract: The emerging drug resistance towards anti-HIV compounds attracted the attention of
scientific community. Development of potent anti-HIV therapy using non-nucleoside reverse
transcriptase inhibitors (NNRTIs) known to be a promising strategy. Although NNRTIs has
great importance in HIV-1 treatment and prevention but their mechanism of action has not yet
been studied in detail. The present review discusses cellular interactions of HIV-1 followed by
the FDA approved anti-HIV-1 chemotherapy. The review highlights the importance of NNRTIs
and their mechanism of action in HIV-1 treatment. Also the current challenges and future
prospective of NNRTIs to prevent HIV-1infection are well addressed so as to propose the
development of novel therapeutic strategies for its treatment.
Key words: HIV-1; non-nucleoside reverse transcriptase inhibitors; mechanism; QSAR;
Computer-aided drug design
Acquired Immunodeficiency Syndrome (AIDS) is a fatal disease caused by infection with the
human immunodeficiency virus (HIV) [1, 2]. In recent years AIDS become a serious global
threat to human health and life. HIV harms body's immune system (T-cells) and hence gradually
destroys the body's ability to fight against various infections as well as certain types of cancer.

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
Reverse transcriptase (RT) an enzyme that catalyzes the transcription of single stranded viral
genomic DNA to double stranded DNA, has been a prominent target in the discovery and
development of therapies for HIV treatment. The first anti-HIV agent approved for clinical use
was a nucleoside reverse transcriptase inhibitor (NRTI) [3]. However, HIV strains with reduced
sensitivity toward NRTI have been isolated from patients receiving prolonged therapy. Further
non-nucleoside HIV-1 RT inhibitors (NNRTI's) were recognized as specific inhibitors of HIV-1.
These compounds induce a conformational change in the three-dimensional structure of the RT
resulted in loss of activity [4]. NNRTIs binding are found to be virus-strain-specific and have
been used in combination with two or three other anti-HIV drugs in Highly Active Antiretroviral
Therapy (HAART).
However, in spite of the remarkable potency of NNRTIs against HIV-1, the rapid clinical
development of NNRTIs is hindered due to more susceptibility toward resistance than any other
classes of antiretroviral drugs. As single-amino-acid change in NNRTI-binding pocket can result
in complete loss of drug activity [5]. Consequently, there is an urgent need to draw attention of
scientific community for the development of new RT inhibitors with an alternative mechanism of
action. The present review focused on the approaches targeted for HIV-1 therapy with special
reference to NNRTI.
Cellular interaction of HIV
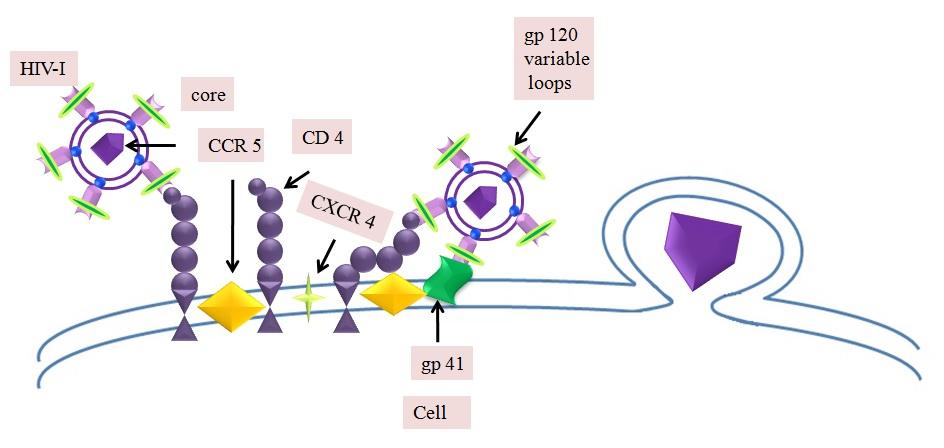
Human CD4+ T lymphocytes, macrophages, microglial, dendritic and Langerhans cells are
considered as potential targets for HIV-1 infection. Endogenous CD4 molecule a 55-KDa protein
member of the IgG super family, is a cell surface receptor expressed on helper T lymphocytes,
monocytes, B-lymphocytes and several other cells. The replication cycle of HIV begins with the
high-affinity binding of the gp120 protein on the host cell surface [6]. This gp120-CD4 complex
interacts with a coreceptor on the cell surface and mediates to complete virus entry into the host
cell. The whole process is assumed to be accomplished with the help of a cytoplasmic peptidyl-
prolyl cis-trans isomerase cyclophilin A (hCyp-18) and appears to be essential for infection by
most HIV-1 strains [7, 8]. The exact role of cyclophilins in HIV-1 infection yet to be fully
explored but studies indicated that virion associated with CyPA may be involved in an early

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
Reverse transcriptase (RT)
HIV-1 RT was discovered in the year 1970 by two independent groups, a Baltimore group and
the Termin and Mizutani group. HIV-1RT, a heterodimer contains subunits of 66 kDa (p66) and
51 kDa (p51). p66 subunit contains two domains, the N-terminal polymerase domain of about
440 residues and the C-terminal RNase H domain with 120 residues. p51 subunit is processed by
proteolytic cleavage of p66 and corresponds to the polymerase domain of the p66 subunit.
Portions of both p51 and the polymerase domain of p66 can be described as a "right hand" which
contains three sub domains: Fingers, palm, and thumb. The connection sub domain connects the
hand of the polymerase domain and RNAse H domain of p66. The connection sub domains and
the palm sub domains comprise of three-stranded β-sheets with -helices on one side whereas
the thumb sub domains include three -helices. There have been 245 structures of HIV-1 RT
deposited in the Protein Database so far.
Fig. 1: Illustrations of cellular interactions of HIV-1 with Human CD4+ T lymphocytes
Anti-HIV chemotherapy
The treatment of the acquired immunodeficiency syndrome (AIDS) is the most challenging
problem worldwide for medical scientist. Anti-HIV compounds that have been approved by FDA
for clinical use in the treatment of AIDS are summarized in Table-1. These compounds fall into
six categories: nucleoside reverse transcriptase inhibitors (NRTIs: zidovudine, didanosine,

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
zalcitabine, stavudine, lamivudine, abacavir and emtricitabine); nucleotide reverse transcriptase
inhibitors (NtRTIs: tenofovir); non-nucleoside reverse transcriptase inhibitors (NNRTIs:
nevirapine, delavirdine, efavirenz and etravirine); protease inhibitors (PIs: saquinavir, ritonavir,
indinavir, nelfinavir, amprenavir, lopinavir, atazanavir, fosamprenavir, tipranavir and darunavir);
cell entry inhibitors [fusion inhibitors (FIs: enfuvirtide) and co-receptor inhibitors (CRIs:
maraviroc)]; and integrase inhibitors (INIs: raltegravir). These compounds should be used in
drug combination regimens (Table 1) to achieve the highest possible benefit, tolerability and
compliance and to diminish the risk of resistance development.
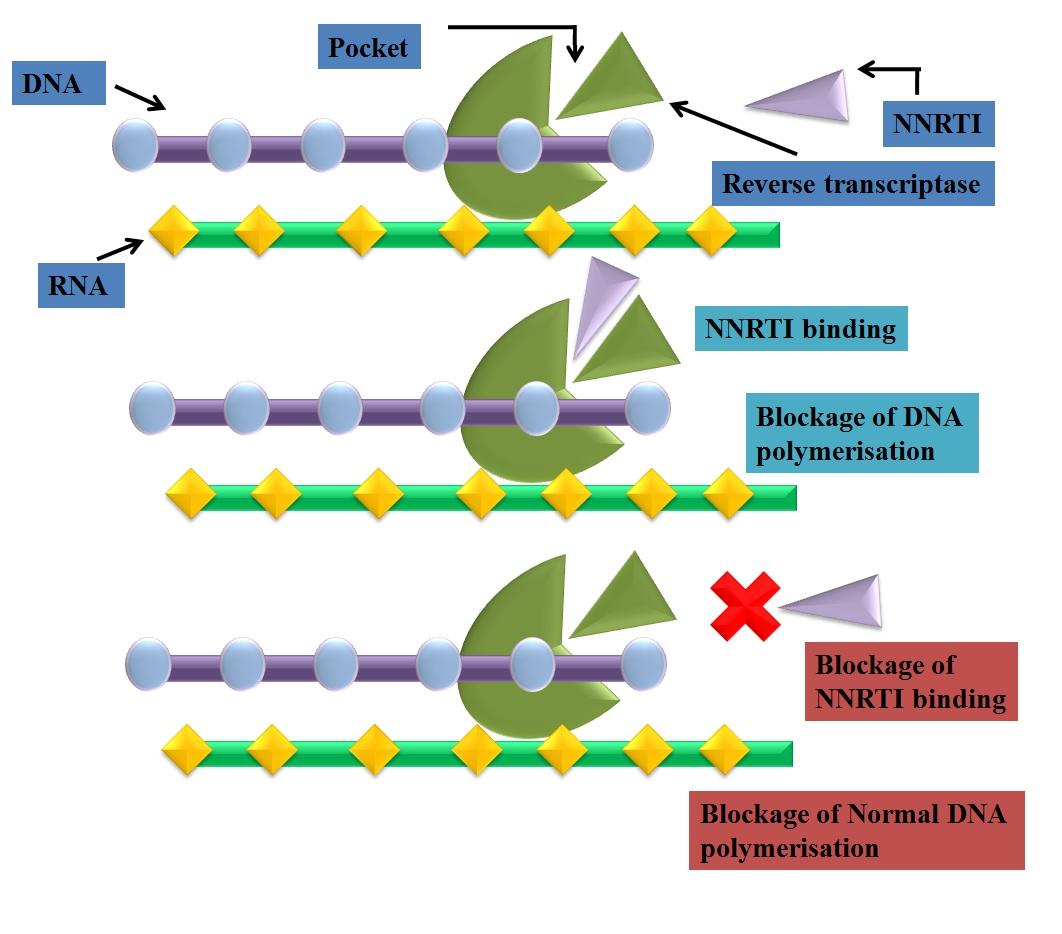
Fig. 3: mechanism of action of NNRTI's

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
Table 1. Anti-HIV compounds approved by FDA in the treatment of AIDS
Nucleoside
Generic name
approval
Inhibitors
Bristol-Myers Squibb
Bristol-Myers Squibb
Tenofovir Disoproxil
Rescriptor Viiv Healthcare
Bristol-Myers Squibb
Inhibitors
(NNRTIs)
Pharmaceuticals, Inc.
Janssen Pharmaceuticals,
Protease
Bristol-Myers Squibb
Inhibitors (PIS)
Janssen Pharmaceuticals,
Merck & Co., Inc
Boehringer Ingelheim
Fusion Inhibitors Enfuvirtide
Hoffmann-La Roche
Entry Inhibitors
Integrase
Inhibitors
Merck & Co., Inc.
Combination
abacavir and lamivudine
HIV Medicines
abacavir, dolutegravir,
and lamivudine abacavir, lamivudine,
and zidovudine efavirenz, emtricitabine,
tenofovir disoproxil

Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
fumarate elvitegravir, cobicistat†,
emtricitabine, and tenofovir disoproxil fumarate emtricitabine, rilpivirine,
and tenofovir disoproxil fumarate emtricitabine and
tenofovir disoproxil fumarate lamivudine and
zidovudine lopinavir and ritonavir
Mechanism of NNRTIs
Non-nucleoside HIV-1 RT inhibitors (NNRTI's) were the first to be recognized as specific
inhibitors of HIV-1. These have been identified by iterative screening using purified viral
enzyme [9] and included a variety of chemical substrates which binds to hydrophobic pocket of
p66 subunit of HIV-1 reverse transcriptase. The non nucleoside reverse transcriptase binding
pocket (NNRTBP) has been found to a close distance from the active catalytic site of HIV-1 RT
and considered as non essential for enzyme function. NNRTIs bind noncompetitively at
NNRTBP and induce a conformational change in the three-dimensional (3D) structure of enzyme
results in loss of enzyme activity [4]. As a result of NNRTI binding, rate of dNTPs incorporation
into synthesizing DNA strand slower down.
Drug Design
Computational methods have developed into very useful tools in facilitating new drug discovery
[10]. By the use of computational methods, it becomes possible to estimate the biological activity
of the candidate molecules even before experimental trials. These methods are simple and non-
expensive and expedite to design molecules with desirable biological activity [11]. Quantitative
structure-activity relationship (QSAR) and docking procedure are two mostly used
computational methods in drug design.
In a study, hologram quantitative structure-activity relationship (HQSAR) was applied to three
different data sets, such as 70 TIBO, 101 HEPT and 125 dipyridodiazepinone derivatives with
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
the inhibitory effect on enzyme activity [12]. Similarly Zhou and Madura (2004) docked 50
TIBO inhibitors and demonstrated that combination of ligand-based and receptor-based
modeling as a powerful approach to build 3D-QSAR models [13]. Another group of researcher
studied quantitative structure-activity relationship (QSAR) analysis for a set of 82 TIBO
using three-layered neural network [14]. Medina-Franco et al. (2004) applied a set of pyridinone
derivatives to study the interactions between pyridinone derivatives and binding pocket of non-
nucleoside reverse transcriptase inhibitor using CoMFA, CoMSIA and docking experimentation
[15]. Also researchers explained the viral resistance to pyridinone derivatives upon mutation of
amino acids Tyr181 and Tyr188. In their other study they designed 44 non-nucleoside HIV-1
reverse transcriptase inhibitors (NNRTIs) of the pyridinone derivative [16] and found pyrazolo
[3,4-d] pyrimidine and phenothiazine as promising novel compounds for NNRTIs. Another class
of HIV-1 non-nucleoside reverse transcriptase (RT) inhibitors includes indolyl aryl sulfones
(IASs) have been studied against wild type and resistant strains (Y181C, the double mutant
K103N-Y181C, and the K103R-V179D-P225H strain). A predictive 3-D QSAR models have
been designed using receptor-based alignment of IASs into the non-nucleoside binding site
(NNBS) of RT [17]. Similarly Duchowicz et al. (2006) studied 154 non-nucleoside reverse
transcriptase inhibitors (NNRTI) of the wild-type HIV-1 virus which considered as the second
generation analogues of Efavirenz [18]. A series of 74 dihydroalkoxybenzyloxopyrimidines
(DABOs) have been studied by comparative molecular field analysis (CoMFA) in order to
design more potent DABO analogues as anti-HIV/AIDS agents [19]. Hu et al. 2009 carried out
molecular modeling of a series of non-nucleoside reverse transcriptase inhibitors (2-amino-6-
arylsulfonylbenzonitriles and their thio and sulfinyl congeners) using comparative molecular
field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA)
approaches [20]. Study revealed, 2-amino-6-arylsulfonylbenzonitriles with bulky and
hydrophobic groups in 3- and 5-position of the B ring showed significant inhibitory activity.
Group of researcher investigated symmetric formimidoester disulfides (DSs) as a new class of
potent non-nucleoside HIV-1 reverse transcriptase (RT) inhibitors using computational strategy
[21]. Liu et al. (2010) discovered and studied a series of 4,6-diamino-1,3,5-triazin-2-ol based
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
NNRTIs compounds using SAR approaches [22]. Samuelea et al. (2010) reported Novel
benzimidazol-2-one non-nucleoside reverse transcriptase inhibitors (NNRTIs) through rational
structure-based molecular modeling and docking approaches [23]. Elena et al. (2011) identified
3,4,5-Trisubstituted-1,2,4-4 H-triazoles (TTs) as a new class of potent non-nucleoside HIV-1
reverse transcriptase (RT) inhibitors [24]. Furthermore they studied 1,3,4,5-tetrasubstituted-
pyrazoles (TPs) as NNRTIs by computational strategy based molecular docking approaches
[25]. Group of researchers conducted a study on a series of 119 NNRTIs, including 1-(2-
hydroxyethoxymethyl)-6-(phenylthio) thymine (HEPT) and dihydroalkoxybenzyloxopyrimidine
(DABO) derivatives using computational approaches [26]. Recently a series of 107 1-[(2-
hydroxyethoxy)-methyl]-6-(phenylthio) thymine (HEPT) as a non-nucleoside reverse
transcriptase inhibitor (NNRTI) has been deliberated [27].
Current challenges
NNRTIs are more susceptible to high-level drug resistance than other classes of antiretroviral
drugs because of a single-amino-acid change in the NNRTI-binding pocket. Due to this reason
the clinical development of NNRTIs is hindered [28]. The amino acids with which the NNRTIs
interact within the NNRTI-binding pocket may be prone to mutate, and this has proven to be the
case for, among others. Most of the NNRTI resistance mutations are found in and around the
NNIBP. K103N and Y181C are the most frequently observed resistance mutations in patients
treated with the approved NNRTIs. When, compared with the ‘older' NNRTIs (e.g. nevirapine),
the ‘newer' NNRTIs such as Etravirine and Rilpivirine first described by Janssen, et al. in 2005,
have been found to retain sufficient activity against the K103N and Y181C reverse transcriptase
(RT) mutants [29]. Other NNRTI resistance mutations that are observed in patients include
L100I, K101E, V106A, V179D, Y188L, G190A, and P236L; these NNRTI resistance mutations
can occur singly, or in combinations. The most promising third generation NNRTIs are proven to
be more effective against HIV strains that carry most common single and double mutations;
however, viral strains carrying multiple NNRTI-resistance mutations can exhibit significant
levels of drug resistance. Currently there are four NNRTI drugs Nevirapine, Delavirdine (first
generation), Efavirenz (second generation), and Etravirine (third generation)) that are approved
for treating HIV-1 infections while several other potent NNRTIs that inhibit HIV-1 at nanomolar
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
concentrations (EC50) are in clinical trials. In recent years, extensive crystallographic, molecular
modeling and biochemical studies have contributed towards understanding NNRTI drug
resistance and the development of better NNRTIs with improved drug profile.
Conclusion and future perspectives
Reverse transcriptase is known as a key enzyme needed for HIV-1 replication. The present
review gives a brief summary of the last decade investigations for designing anti-HIV agents
(NNRTIs) using computer-aided approaches. It has been found that NNRTIs could play an
important role to design a promising therapy for HIV treatment. Also the development of QSAR
based drug designing models with a novel mechanism of actions could improve the pace of new
anti-HIV drug inventions. It is essential to study and Understanding the molecular mechanism of
NNRTI inhibition to overcome the drug-resistance mutations in RT. Further scientific
communities should be more focused on highly specific NNRTI inhibitors with low toxicities
and minimal side effects.
Conflict of Interest
There is no potential conflict of interest with reference to the current manuscript. All the authors
have read the manuscript and agreed to submit the same for publication.
The authors would like to acknowledge Mata Gujri Collage (Fatehgarh Sahib) for providing the
requisite facilities to carry out this work.
References
1. Gallo RC, Sarin PS and Gelmann EP (1983). Isolation of human T-cell leukemia virus in
acquired immune deficiency syndrome (AIDS). Science 220: 865–867.
2. Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C,
Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W and Montagnier L (1983).
Isolation of a T-lymphotropic retrovirus 66 from a patient at risk for acquired immune
deficiency syndrome (AIDS). Science 220: 868-871.
3. Fischl MA, Richman DD and Grieco MH (1987). The efficacy of azidothymidine (AZT)
in the treatment of patients with AIDS and AIDS-related complex: A double-blind,
placebo-controlled trial. New Engl J Med 317:185-191.
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
4. Spence RA, Kati WM, Anderson KS and Johnson KA (1995). Mechanism of inhibition
of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267: 988-993.
5. Maga G, Radi M, Gerard MA, Botta M and Ennifar E (2010). HIV-1 RT inhibitors with a
novel mechanism of action: NNRTIs that compete with the nucleotide substrate. Viruses
6. Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF and Weiss RA
(1984). The CD4 (T4) antigen is an essential component of the receptor for the AIDS
retrovirus. Nature 312: 763-767.
7. Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP (1993). Human
immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73: 1067-
8. Luban J (1996). Absconding with the chaperone: essential cyclophilin-Gag interaction in
HIV-1 virions. Cell 87: 1157-1159.
9. Pauwels R, Andries K and Desmyter J (1990). Potent and selective inhibition of HIV-1
replication in vitro by a novel series of TIBO derivatives. Nature 343: 470-474.
10. Kuntz ID (1992). Structure-based strategies for drug design and discovery. Science 257:
11. Lorber DM (1999).Computational drug design. Chem Biol 6:R227-28.
12. Pungpo P, Hannongbua S and Wolschann P (2003). Hologram quantitative structure-
activity relationships investigations of non-nucleoside reverse transcriptase inhibitors.
Curr Med Chem 10(17): 1661-1677.
13. Zhou Z and Madura JD (2004). CoMFA 3D-QSAR analysis of HIV-1 RT nonnucleoside
inhibitors, TIBO derivatives based on docking conformation and alignment. J Chem Inf
Comput Sci 44(6): 2167-2178.
14. Douali L, Villemin D and Cherqaoui1 D (2004). Exploring QSAR of non-nucleoside
reverse transcriptase inhibitors by neural networks: TIBO Derivatives. Int J Mol Sci 5:
15. Medina-Franco JL, Rodríguez-Morales S, Juárez-Gordiano C, Hernández-Campos A and
Castillo R (2004). Docking-based CoMFA and CoMSIA studies of non-nucleoside
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
reverse transcriptase inhibitors of the pyridinone derivative type. J Comput Aided Mol
Des 18(5): 345-360.
16. Medina-Franco JL, Golbraikh A, Oloff S, Castillo R and Tropsha A (2005). Quantitative
structure-activity relationship analysis of pyridinone HIV-1 reverse transcriptase
inhibitors using the k nearest neighbor method and QSAR-based database mining. J
Comput Aided Mol Des19 (4): 229-242.
17. Ragno R, Artico M, de Martino G, La Regina G, Coluccia A, Di Pasquali A and Silvestri
R (2005). Docking and 3-D QSAR studies on indolyl aryl sulfones. Binding mode
exploration at the HIV-1 reverse transcriptase non-nucleoside binding site and design of
highly active N-(2-hydroxyethyl) carboxamide and N-(2-hydroxyethyl) carbohydrazide
derivatives. J Med Chem 48(1): 213-223.
18. Duchowicz PR, Fernández M, Caballero J, Castro EA and Fernández FM (2006). QSAR
for non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg Med Chem 14(17):
19. de Brito MA, Rodrigues CR, Cirino JJ, de Alencastro RB, Castro HC and Albuquerque
MG (2008). 3D-QSAR CoMFA of a series of DABO derivatives as HIV-1 reverse
transcriptase non-nucleoside inhibitors. J Chem Inf Model 48(8): 1706-1715.
20. Hu R, Barbault F, Delamar M and Zhang R (2009). Receptor- and ligand-based 3D-
QSAR study for a series of non-nucleoside HIV-1 reverse transcriptase inhibitors.
Bioorg. Med Chem.17(6): 2400-2409.
21. Cichero E, Cesarini S, Spallarossa A, Mosti L and Fossa P (2009). Computational studies
of the binding mode and 3D-QSAR analyses of symmetric formimidoester disulfides: a
new class of non-nucleoside HIV-1 reverse transcriptase inhibitor. J Mol Model 15(4):
22. Liu B, Lee Y, Zou J, Michael Petrassi H, Joseph RW, Chao W, Michelotti EL,
Bukhtiyarova M, Springman EB and Dorsey BD (2010). Discovery and SAR of a series
of 4,6-diamino-1,3,5-triazin-2-ol as novel non-nucleoside reverse transcriptase inhibitors
of HIV-1. Bioorg Med Chem Lett 20:6592–6596.
Journal of Biological and
Chemical Sciences (JBCS)
Sharma et al. (2014) vol.1, no. 1, 1-12
23. Samuelea A, Crespana E, Vitellaroa S, Monforteb AM, Logotetab P, Chimirrib A and
Magaa G (2010). Slow binding–tight binding interaction between benzimidazol-2-one
inhibitors and HIV-1 reverse transcriptase containing the lysine 103 to asparagine
mutation. Antiviral Res 86: 268-275.
24. Elena C, Laura B and Paola F (2011). 3,4,5-Trisubstituted-1,2,4-4 H-triazoles as WT and
Y188L mutant HIV-1 non-nucleoside reverse transcriptase inhibitors: docking-based
CoMFA and CoMSIA analyses. J Mol Model 17: 1537-1550.
25. Elena C, Laura B and Paola F (2012). Docking-based 3D-QSAR analyses of pyrazole
derivatives as HIV-1 non-nucleoside reverse transcriptase inhibitors J Mol Model 18:
26. Mao Y, Li Yan, Hao M, Zhang S and Ai C (2012). Docking, molecular dynamics and
quantitative structure-activity relationship studies for HEPTs and DABOs as HIV-1
reverse transcriptase inhibitors. J Mol Model 18: 2185-2198.
27. Toropovaa AP, Toropova AA, Veselinovićb JB, Miljkovićb FN and Veselinovićb AM
(2014). QSAR models for HEPT derivates as NNRTI inhibitors based on Monte Carlo
method. Eur J Med Chem77: 298–305.
28. Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD,
Bonhoeffer S, Nowak MA and Hahn BH (1995). Viral dynamics in human
immunodeficiency virus type 1 infection. Nature 373(6510): 117–122.
29. Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers
M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Béthune MP, Pauwels
R, Das K, Clark AD Jr, Frenkel YV, Hughes SH, Medaer B, de Knaep F, Bohets H, de
Clerck F, Lampo A, Williams P and Stoffels P (2005). In search of a novel anti-HIV
drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-
2,6 dimethylphenyl]amino]- 2-pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J
Med Chem 48(6): 1901–1909.
Source: http://www.scienceindoors.com/public/vol1/MS-JBCS-D-006-14.pdf
Das genial einfache Abnehmprogramm Schlank über Nacht! Es klingt wie ein Wunder für al e Übergewichtigen auf dieser Welt: Abends dick und fett ins Bett und morgens rank und schlank wieder raus! Ganz so schnell geht es leider nicht, aber ich verspreche Ihnen, dass es funktioniert: Sie können im Schlaf ungewollte Fettpolster verlieren! Mit ein
VISTAS IN GEOLOGICAL RESEARCH (ISBN: 81-900907-0-4) Special Publication in Geology (14), January 2016, pp.157 - 164 A Palynological Investigation of Quaternary Sediment Core from Cauvery Delta, Tamilnadu, India MOHAPATRA , A. KRISHNAMURTHY , P. SRINIVASAN , P. SINGH , P. P. DAS *1Department of Geology, Raveneshaw University, Cuttack - 753003, India















