Untitled
Microbiology (2007), 153, 1394–1404
ScbA from Streptomyces coelicolor A3(2) hashomology to fatty acid synthases and is able tosynthesize c-butyrolactones
Nai-Hua Hsiao,13 Johannes So¨ding,2 Dirk Linke,2 Corinna Lange,3Christian Hertweck,3 Wolfgang Wohlleben1 and Eriko Takano13
Mikrobiologie/Biotechnologie, Eberhard-Karls-Universita¨t Tu¨bingen, Auf der Morgenstelle 28,
72076 Tu¨bingen, Germany
2Max-Planck-Institut fu¨r Entwicklungsbiologie, Spemannstr. 35, 72076 Tu¨bingen, Germany
3Leibniz Institute for Natural Product Research and Infection Biology, HKI, Beutenbergstr. 11a,
07745 Jena, Germany
c-Butyrolactones play an important role in the regulation of antibiotic production and differentiationin Streptomyces. However the biosynthetic pathway for these small molecules has not yet beendetermined, and in vitro synthesis has not been reported. The function of the AfsA family of proteins,originally proposed to catalyse c-butyrolactone synthesis, has been in debate. To clarify the functionof the AfsA family, and to understand the synthesis of the c-butyrolactones, we performed insilico analysis of this protein family. AfsA proteins consist of two divergent domains, each of whichhas similarity to the fatty acid synthesis enzymes FabA and FabZ. The two predicted active sites in
Received 18 November 2006
ScbA, which is the AfsA orthologue found in Streptomyces coelicolor, were mutated, and
c-butyrolactone biosynthesis was abolished in all four constructed mutants, strongly suggesting
Accepted 4 January 2007
that ScbA has enzymic activity.
virginiae butanolides (VBs) isolated from Streptomycesvirginiae; this synthesis is thought to involve a condensation
Bacterial signalling molecules have been shown to be key
of the glycerol derivative dihydroxyacetone and 7-methyl-3-
regulators of differentiation, antibiotic production, plasmid
oxo-octanoyl CoA to form a 7-methyl-3-oxo-octanoic acid.
conjugation, biofilm formation and pathogenicity (see
In the presence of NADH, this is then converted into 6-
reviews: Camilli & Bassler, 2006; Vendeville et al., 2005;
dehydroVB A, and further to VB A in the presence of
Venturi, 2006). c-Butyrolactones are produced by the Gram-
NADPH (Sakuda et al., 1990, 1992, 1993). However, in vitro
positive soil-dwelling bacteria Streptomyces, and they were
synthesis proving this biosynthesis pathway has not been
among the first signalling molecules to be identified
reported. Recently, Shikura et al. (2002) identified a
(Khokhlov et al., 1967). The first and the most well-
stereospecific reductase essential for the last step in VB
characterized c-butyrolactone is A-factor from Streptomyces
griseus, and it is required in nanomolar concentrationsfor antibiotic (streptomycin) production and sporulation.
A putative A-factor biosynthetic gene, afsA, has been cloned
Today, several c-butyrolactones are known, including the
from S. griseus (Horinouchi et al., 1985), and a number of
Streptomyces coelicolor butyrolactones (SCBs), which regulate
homologues of AfsA have subsequently been identified from
actinorhodin and undecylprodigiosin, and the recently
several streptomycetes (see Takano, 2006, for a review).
identified polyketide synthase biosynthesis cluster Cpk in
However, in vitro A-factor synthesis has not yet been
S. coelicolor (see Takano, 2006, for a review; Yamada, 1999).
demonstrated, and little is known about how the synthesis is
The first c-butyrolactone biosynthesis analysis was that of
controlled. Cloning of afsA on a multi-copy plasmid leads toprecocious production of streptomycin in S. griseus, and
synthesis of A-factor in several Streptomyces species that
Present address: Department of Microbial Physiology, GBB, University
of Groningen, Kerklaan 30, 9751NN, Haren, The Netherlands.
normally do not produce it (Horinouchi et al., 1985).
Expression of afsA in Escherichia coli also results in
Full details of the construction of ScbA mutants and further MS dataare available as supplementary data with the online version of this
production of biologically active c-butyrolactones; this
expression is reduced in the presence of cerulenin, a fatty
Abbreviations: SCB, Streptomyces coelicolor butyrolactones; VB,
acid synthesis inhibitor (Ando et al., 1997). From these
results, and those obtained by Sakuda et al. (1990, 1992)
2006/004432 G 2007 SGM
Printed in Great Britain
ScbA active sites are similar to FabA/Z
outlined above, it has been concluded that AfsA on its own
agar (Takano et al., 2001) was used for c-butyrolactone bioassays.
can produce A-factor from a glycerol derivative, and an a-
MS agar (Kieser et al., 2000) was used to make spore suspensions,
keto acid derived from fatty acid biosynthesis.
and for plating out conjugations with E. coli ET12567 containing theRP4-derivative pUZ8002 (Flett et al., 1997). R2 (Kieser et al., 2000)
A contrasting view arose from studies of the gene barX. barX is
and R2YE (Kieser et al., 2000) were used to observe morphologicaldifferentiation and antibiotic production of mutants.
an afsA homologue from S. virginiae that is located adjacent tobarA, which encodes the VB-binding protein. A barX-deletion
Primers, PCR and DNA sequencing. All primers used in this
mutant produces less virginiamycin and VBs, but the addition
work are listed in Table 2. Amplification of DNA by PCR was done
of VBs to the mutant does not restore virginiamycin pro-
with Taq polymerase or ProofStart polymerase (Qiagen). E. coli
duction (Kawachi et al., 2000), in contrast to the situation
colony PCR was carried out by mixing a small amount of cellspicked from a single colony directly into the PCR reaction pre-mix-
reported for the afsA mutant in S. griseus (Hara & Beppu,
ture. DNA was sequenced by MWG.
1982). Furthermore, BarX enhances the stability of theinteraction between BarA and its cognate barB (barA homo-
DNA and RNA manipulations. Plasmid DNA isolation, restric-
logue) promoter by a protein–protein interaction that leads to
tions and cloning experiments were carried out as described bySambrook et al. (1989). Streptomyces genomic DNA was isolated
repression of barB expression. Thus, the authors conclude that
according to Leblond et al. (1996). RNA was isolated as described by
BarX does not possess enzymic activity analogous to AfsA, but
Strauch et al. (1991).
that it may be involved in the regulation of VB synthesis.
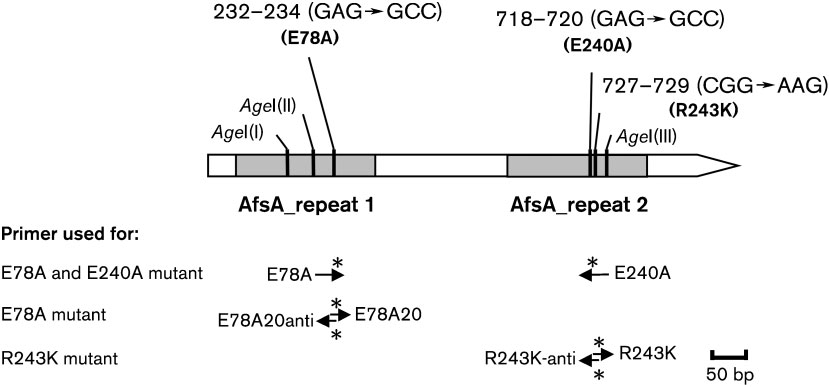
Construction of ScbA mutants. The conserved glutamates E78
scbA is the afsA homologue in S. coelicolor, which is
and E240 of ScbA were mutated to alanine, and the conserved argi-
genetically the most well-characterized streptomycete, and
nine R243 was mutated to lysine by site-directed mutagenesis using
has a completely sequenced genome. scbA is located
SLIM (Chiu et al., 2004), to yield pTE106, pTE108, pTE104 andpTE110. For full details of the construction of ScbA mutants, see the
divergently from the c-butyrolactone-binding protein
supplementary data available with the online version of this paper.
determinant scbR (Takano et al., 2001), thus exhibitingthe same gene organization as S. virginiae (see Takano, 2006,
Introduction of the mutation into Streptomyces. pTE104,
for a review). A deletion mutant of scbA does not produce
pTE106, pTE108 and pTE110 were introduced into the methylation-
any c-butyrolactones with antibiotic stimulatory activity,
deficient E. coli strain ET12567 containing the RP4-derivativepUZ8002 (Paget et al., 1999), and transferred to S. coelicolor M751
and expression of scbA is absent in both scbA- and scbR-
by conjugation (Flett et al., 1997); single-crossover exconjugants
deletion mutants. While the expression of scbR is induced by
were selected on MS containing apramycin and nalidixic acid to
addition of SCB1, a c-butyrolactone, to the scbA mutant,
obtain transconjugants M751 : : pTE104, M751 : : pTE106, M751 : :
scbA transcription is not inducible under the same
pTE108 and M751 : : pTE110, respectively. The genomic DNA of
conditions (Takano et al., 2001). Thus, some direct role
each strain was isolated, and plasmid integration was confirmed by
for ScbA in transcriptional activation of its own gene
PCR with primers JGatt B1-fwd and JGatt Pint-rev (Table 2), which
appears probable, suggesting a possible regulatory function
gave an amplified product of 0.8 kb, corresponding to the insertedregion (data not shown).
for ScbA. Production of SCBs is detected at transition tostationary phase, and this also coincides with the transcrip-
RT-PCR was conducted using RNA isolated from
tion of scbA (Takano et al., 2001).
M751 : : pSET152 (negative control: mutant complemented withvector), M751 : : pIJ6147 (positive control: mutant complemented
To clarify these conflicting results, and to understand the
with wt scbA), M751 : : pTE104, M751 : : pTE106, M751 : : pTE108
function of ScbA and the other members of the AfsA family,
and M751 : : pTE110 grown in SMM liquid medium for 24 h (seeFig. 3a). A 2 mg quantity of RNA was used to synthesize the cDNA,
we undertook an in silico analysis of the protein sequences.
and 27 ng of this cDNA was used as the template for the PCR reac-
From these analyses, a number of conserved residues that are
tion, as previously reported, using primers for scbA, hrdB, redQ,
likely to participate in an enzymic function of ScbA were
redD, actII-ORF4, cpkO, cpkE and actIII, except that the PCR condi-
identified. Point mutations in these positions were created,
tions in the present study were 96 uC for 5 min, then 30 cycles of
and the resulting S. coelicolor strains were tested for the
96 uC for 40 s, 58 uC for 40 s, 72 uC for 40 s, and extension at 72 uC
ability to produce c-butyrolactones with antibiotic stimu-
for 7 min (Takano et al., 2005).
latory activity. All the point mutations tested resulted in loss
HPLC-MS monitoring of c-butyrolactone formation. Extracts
of antibiotic stimulatory function, indicating that ScbA is
from SMMS-grown M751 : : pSET152, M751 : : pIJ6147, M751 : :
indeed an enzyme involved in c-butyrolactone synthesis,
pTE104, M751 : : pTE106, M751 : : pTE108 and M751 : : pTE110,
and that it is structurally and functionally related to bacterial
SMMS (medium only), and the standard (SCB1), were dissolved in
fatty acid synthesis enzymes.
methanol. All experiments were done using an Agilent 1000 SeriesLC/MSD ion trap system. The system was operated with the electro-spray ionization source in the positive mode. HPLC conditions wereas follows: the column was an Agilent Zorbax Eclipse XDB C8
(5 mm, 150
64.6 mm); the mobile phase consisted of A (water) andB (methanol); the gradient elution started with 10 % at 1 min to
Bacterial strains, plasmids and growth conditions. Strains and
100 % at 17 min, and was then kept at 100 % for 5 min, flow rate
plasmids used in this study are listed in Table 1. Streptomyces strains
1.0 ml min21. MS conditions: dry temperature 350 uC, nebulizer
were manipulated as described by Kieser et al. (2000). E. coli was
60 p.s.i., dry gas 11 l min21, ion mode positive (MS and MS2).
grown and transformed according to Sambrook et al. (1989). SMM(Takano et al., 2001) was used for RNA isolation, isolation of c-
Other methods. c-Butyrolactones were isolated and the bioassay
butyrolactones, and antibiotic production determination. SMMS
was conducted as described previously by Takano et al. (2001).
N.-H. Hsiao and others
Table 1. Strains and plasmids used in this study
Strain or plasmid
Source or reference
S. coelicolor A3(2)M145
Wild-type (genome sequenced)
Kieser et al. (2000)
In-frame deletion of scbA
Takano et al. (2001)
pSET integrated into H145
pSET152 integrated in M751
pIJ6147 (full-length scbA) integrated in M751
pTE104 (Glu-Ala scbA mutant) integrated in M751
pTE106 (E78A scbA mutant) integrated in M751
pTE108 (E240A scbA mutant) integrated in M751
pTE110 (Arg-Lys scbA mutant) integrated in M751
General-purpose cloning strain
Sambrook et al. (1989)
Strain used for conjugation between E. coli and Streptomyces
MacNeil et al. (1992)
Non-replicative conjugative cloning plasmid
Bierman et al. (1992)
Non-transmissible transfer plasmid
Flett et al. (1997)
PCR product cloning vector
Expression vector
scbA within pGEM-T-Easy vector
Takano et al. (2001)
scbA within pSET152 vector
Takano et al. (2001)
1 kb scbA PCR product cloned into pGEM-T Easy
562 bp mutated scbA PCR product cloned into pGEM-T Easy
AgeI scbA fragment of pTE101 cloned into pIJ6143 (lost 49 nt)
pTE102 derivative containing full length of scbA (with 49 nt)
EcoRI scbA fragment of pTE103 cloned into pSET152
pIJ6143 derivative containing E78A mutated scbA
EcoRI DNA fragment of pTE105 subcloned into pSET152
NdeI/BbpI DNA fragment from pTE103 subcloned into pIJ6143
EcoRI scbA fragment of pTE107 cloned into pSET152
pIJ6143 derivative containing Arg-Lys mutated scbA
EcoRI DNA fragment of pTE105 subcloned into pSET152
M145 : : pSET152, M751 : : pSET152, M751 : : pIJ6147, M751 : : pTE104,
hidden Markov models (So¨ding et al., 2005), revealed that
M751 : : pTE106, M751 : : pTE108 and M751 : : pTE110 were grown in
ScbA consists of two domains homologous to the super-
SMM liquid or on SMMS solid medium, and ethyl acetate extracts
family of thioesterase/thiol ester dehydrase-isomerases
were made from stationary-phase culture supernatants at OD450 1.2
defined in the SCOP database (Murzin et al., 1995). Each
(the last time point in Fig. 3a). Equal amounts of the supernatantextracts were spotted onto confluent lawns of the indicator M145.
of these domains contains the described AfsA repeat as their
After 48 h incubation, the antibiotic stimulatory activity was deter-
most conserved part. Two members of the thioesterase/thiol
mined (see Fig. 3b). Antibiotic production assay was conducted using
ester dehydrase-isomerase superfamily, namely FabA (b-
1 ml samples of cells grown in 50 ml SMM, as described by Strauch
hydroxydecanoyl thiol ester dehydrase) and FabZ [(3R)-
et al. (1991).
hydroxymyristoyl ACP dehydrase], have several conservedactive site positions that also appear in both domains of ScbA
(see below). Moreover, FabA and FabZ are functional ashomodimers whose two active sites are located at the dimer
ScbA has active sites that are similar to FabA
interface (Leesong et al., 1996; Kostrewa et al., 2005). This
strongly suggests a similar spatial arrangement of the twodomains in ScbA. Although the overall sequence similarity of
ScbA contains two AfsA repeats according to Pfam
ScbA to FabA and FabZ is weak (15 and 13 % amino acid
(Pfam03756) (Sonnhammer et al., 1998). These repeats
identity, respectively), the similarity in the region around the
show no similarity to any other proteins in the databases
FabA/FabZ active site is pronounced, and, in particular, the
when using standard BLAST or PSI-BLAST searches (Altschul
hydrophobicity pattern is well conserved (Fig. 1).
et al., 1997). A sequence search of all proteins with knownstructure using the advanced remote homology detection
The active site in FabA and FabZ is formed between the long
tool HHpred, which is based on pairwise comparison of
a-helix of one subunit, and a loop that is N-terminal to the
ScbA active sites are similar to FabA/Z
Table 2. Primers used in this work
Sequence (5§R3§)
*The AgeI site is underlined, and nucleotide changes are shown in upper-case type.
DThe insertion sequence is shown in bold upper-case type.
dThe nucleotide changes are shown in upper-case type.
equivalent helix of the other subunit. The residues involved
well conserved in both domains of ScbA (R81, R243), and
in catalysis are a conserved aspartate/glutamate located in
that is situated just before the conserved glutamine, but not
the helix, and a conserved histidine present in the loop
present in FabA or FabZ.
region that forms the catalytic diad. A conserved glutamineforms a hydrogen bond with the carboxyl side chain of theconserved glutamate/aspartate residue, and stabilizes its
Mutagenesis of the predicted active-site
position (Leesong et al., 1996; Kimber et al., 2004; Kostrewa
residues in ScbA: E78, E240 and R243
et al., 2005). In ScbA, the glutamates (E78 and E240) and
To test the prediction that these homologous amino acids
glutamines (Q82 and Q244) are perfectly conserved in both
are involved in forming the active site of ScbA, the conserved
domains, while the histidine residue is present only in the C-
E78 and E240 were mutated to alanine, and the conserved
terminal domain. In other homologues, the C-terminal
R243 was mutated to lysine, in the AfsA repeat domains. E78
histidine is replaced by an arginine (H226R) (Fig. 1). This
and E240 were mutated to alanine separately to yield
may be related to, or the functional difference between ScbA
pTE106 (E78A) and pTE108 (E240A), respectively, and
and FabA/FabZ may be due to, the arginine residue that is
together to yield pTE104 (EA). The conserved R243 was
Fig. 1. ScbA consists of two divergent, duplicated domains that are remotely, but significantly, homologous to FabA and FabZ, and share most of their key catalyticresidues. The multiple alignment of the N- and C-terminal domains of ScbA, FabA and FabZ, and their homologues, was generated by HHsearch. Amino acids are colouredaccording to their physical–chemical properties: positive, blue; negative, red; polar, magenta; small, black; aromatic, dark green; non-polar, light green; and proline, orange.
Conserved amino acid residues are boxed. Residues in b-sheets and a-helices are marked with E and H, respectively. The black squares indicate the active sites of FabAand FabZ. The asterisk, hash and black circle indicate the probable functional residues E78, E240 and E243, respectively, that were mutated in our experiments.
ScbA active sites are similar to FabA/Z
Fig. 2. Schematic map of amino acid exchanges made in ScbA. scbA, represented as an open arrow, has two AfsA repeatdomains (filled boxes) and three AgeI sites (vertical black lines). Mutated DNA sites at positions 232–234 (E78A), 718–720(E240A) and 727–729 (R243K) are indicated by vertical black lines. The primers used for mutagenesis are shown by arrows,and the mutated sites are labelled by asterisks.
mutated to yield pTE110 (R243K) (Methods; Figs 1 and 2).
the loss of two molecules of water (see supplementary Figs S1
These constructs were then introduced into the scbA
and S2 available with the online version of this paper).
deletion mutant M751 by conjugation, to give strainsM751 : : pTE104, M751 : : pTE106, M751 : : pTE108 and
To exclude any possibility that the mutated scbA constructs
M751 : : pTE110. The original promoter was retained in all
were not being correctly expressed, RT-PCR was conducted,
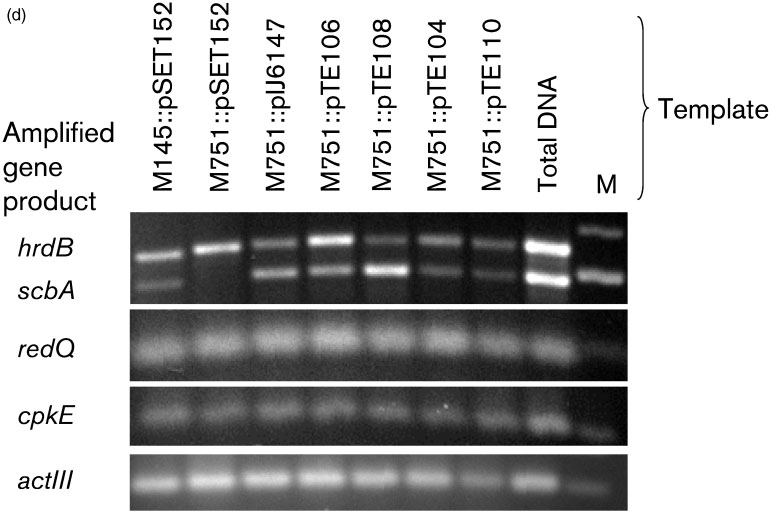
as described in Methods. The expression of hrdB encodingthe major sigma factor for S. coelicolor was readily detectedin all samples (Fig. 3d), while the scbA transcript was
All mutated ScbA strains lost the ability to
detected in all the samples except M751 : : pSET152, which is
produce c-butyrolactones, even though the
the scbA deletion mutant with an integrated vector only
mutated genes were expressed
(Fig. 3d). No amplified products were detected using RNA
The ability of the constructed mutants to produce SCB1 c-
as a template, which suggests that there was no DNA
butyrolactone was tested by bioassay, looking for the ability
contamination of the RNA samples (data not shown). This
of ethyl acetate extracts isolated from stationary-phase
suggests that scbA was expressed in the mutants, and that the
cultures of the strains to induce pigmented antibiotic
mutations, and not the loss of transcription, led to the loss of
production in the indicator strain M145 (as detailed in
bioactive c-butyrolactone production.
Methods). The results are presented in Fig. 3(b). Asreported previously, an extract from the positive control
To assess the effect of the mutations on growth and
strain M751 : : pIJ6147 stimulated pigmentation; however, it
antibiotic production, M751 : : pTE104, M751 : : pTE106,
needed extracts that were twofold more concentrated than
M751 : : pTE108 and M751 : : pTE110 were grown on several
that of M145 : : pSET152. Extracts from M751 : : pTE104,
different solid media (R2, SMMS, R2YE and MS) in
M751 : : pTE106, M751 : : pTE108 and M751 : : pTE110
triplicate, but they showed only slight differences compared
showed no antibiotic stimulatory activity, even when
with wt : : pSET152 or M751 : : pSET152 (data not shown;
using a twofold excess of supernatant extract compared
see Discussion). The mutants were also grown on SMM
with the positive control (Fig. 3b). This result indicates that
liquid media at 30 uC in triplicate, and, after 18 h
the amino acid replacements (E78A, E240A and R243K) in
incubation, samples were taken at four time points (every
these two putative active sites leads to an inability to
2 h) and after 34 h at OD450. The level of antibiotic
produce active c-butyrolactones.
production of undecylprodigiosin and actinorhodin wasdifferent at each time point, and no conclusion could be
These results were corroborated by analyses of the metabolic
made from these results (data not shown). RT-PCR using
profiles by HPLC-MS, using a synthetic reference of
the undecylprodigiosin-pathway-specific activator RedD,
SCB1. As shown in Fig. 3(c), only M145 : : pSET152 and
and a representative biosynthesis gene, redQ, the actinor-
M751 : : pIJ6147 were capable of producing SCB1, while no
hodin-pathway-specific activator actII-ORF4, and a repre-
related c-butyrolactones were detected in the mutants. MS
sentative biosynthesis gene, actIII, the Cpk pathway-specific
revealed the characteristic fragmentation that is initiated with
activator CpkO, and a representative biosynthesis gene,
N.-H. Hsiao and others
ScbA active sites are similar to FabA/Z
Fig. 3. Amino acid exchange mutants do not produce bioactive c-butyrolactones. (a) Growth curves of parent (&), andmutants M751 : : pSET152 ($), M751 : : pIJ6147 (m), M751 : : pTE104 (.), M751 : : pTE106 (X), M751 : : pTE108 (b) andM751 : : pTE110 (c). Strains were grown in SMM liquid medium at 30 6C. At time points 18, 20, 22, 24 and 34 h, sampleswere collected to measure OD450. (b) c-Butyrolactone bioassays. Ethyl acetate extracts from M145 : : pSET152,M751 : : pSET152, M751 : : pIJ6147, M751 : : pTE104, M751 : : pTE106, M751 : : pTE108 and M751 : : pTE110 culturesupernatants were spotted onto confluent lawns of M145 on SMMS, and incubated at 30 6C for 48 h. Antibiotic stimulatoryactivity (purple halo) indicates that the strain produces active c-butyrolactones. Chemically synthesized SCB1 (0.25 ng) wasused as the positive control, and methanol and ethyl acetate were spotted as negative controls. (c) HPLC-MS profiles ofextracts from wild-type and mutants. Displayed is single ion monitoring at m/z 245. Ethyl acetate extracts from SMMS-grownM145 : : pSET152,
M751 : : pSET152,
M751 : : pIJ6147,
M751 : : pTE110, SMMS (medium only), and chemically synthesized SCB1, were analysed by HPLC. The peak correspondingto SCB1 was seen in samples SCB1, M145 : : pSET152 and M751 : : pIJ6147, and is shown by an arrow. Sample names areindicated in the trace box. (d) Gene expression in the constructed mutants of scbA. RT-PCR using cDNA synthesized fromRNA
M145 : : pSET152,
M751 : : pSET152,
M145 : : pIJ6147,
M751 : : pTE108 and M751 : : pTE110 was conducted. Amplified products were run on an agarose gel, and the amplifiedgene products are indicated on the left. The expected sizes of the amplified products were: hrdB, 550 bp; scbA, 480 bp;redQ, 80 bp; cpkE, 95 bp; and actIII, 97 bp. All PCR was performed at 30 cycles. The template used for the positive controlfor all primers was M145 total DNA. M, 100 bp DNA ladder.
cpkE, was conducted in triplicate to determine the effect of
Our results point strongly to a role for ScbA in the
the mutation on the expression of these genes. The
biosynthesis of SCB1.
expression levels of the three biosynthesis genes were similarin all the strains tested (Fig. 3d). The three activators
Standard BLAST analysis of the AfsA family of proteins has
showed varied expression levels (data not shown), which
not revealed any similarity to other proteins in the database
may correspond to the growth phase in which the RNA was
since the sequencing of AfsA in 1985. By using the novel
isolated (see Discussion).
software HHpred (So¨ding et al., 2005), we were able todetermine, for what we believe is the first time, that ScbA hasactive sites similar to fatty acid synthesis genes. The synthesisof fatty acids is performed by the type II fatty acid
biosynthesis pathway in eubacteria. There are four steps in
The precise mechanism of ScbA/AfsA homologues in c-
the elongation cycle, which extends two carbons per cycle. In
butyrolactone synthesis has been under considerable debate
E. coli, the dehydration of the b-hydroxyl-ACP is performed
for the last decade. Moreover, the exact biosynthetic
by FabA or FabZ. Unsaturated fatty acids are produced by
pathway for the c-butyrolactones is unknown, despite
the isomerase FabM, utilizing the fatty acid intermediate
their importance in the regulation of antibiotic production.
produced by FabZ, or directly by FabA, which has been
N.-H. Hsiao and others
shown to have an additional trans-2- to cis-3-decenoyl-ACPisomerase activity (Heath & Rock, 1996; Brock et al., 1967).
FabZ, on the other hand, does not exhibit an isomeraseactivity, and is involved in both saturated and unsaturatedfatty acid elongation. FabA is mainly found in Gram-negative bacteria that produce unsaturated fatty acids, whileFabZ is found in most bacteria (Kimber et al., 2004). The 3Dstructures and the active sites have been determined for bothFabA and FabZ. The active site residues determined frommutational studies are H70 and D84 for E. coli FabA(Leesong et al., 1996), and H133 and E147 for Plasmodiumfalciparum FabZ (Kostrewa et al., 2005).
We chose to mutate the conserved glutamates in the twoAfsA repeat domains of ScbA. The other active site residues,histidine, in the fatty acid synthases was not conserved in theN-terminal domain of ScbA, but only in the C-terminalregion, so this histidine was not mutated. The mutated C-terminal R243, on the other hand, was not conserved in thefatty acid synthase, but only in the AfsA repeat domains. Themutagenesis of either one or both conserved glutamateresidues, or the arginine, abolished the production of c-butyrolactones with antibiotic stimulatory activity. Thisstrongly suggests that these amino acids are important for
Fig. 4. Proposed structural model of ScbA. Modelled structure
ScbA activity, indicating the HHpred prediction was indeed
of ScbA deduced from the homology to FabA and FabZ, with
correct, and that ScbA is indeed an enzyme homologous to
their duplicated ‘hot dog' fold, showing the two active catalytic
FabA and FabZ. The exact enzymic role of ScbA will need
sites that presumably bind fatty acid derivatives. The classic
further biochemical analysis, but from the similarity of the
inhibitor of FabZ is 3-decanoyl-N-acetylcysteamine, and this is
active sites to FabA and FabZ, a dehydrase activity is
modelled into the structure (green spacefill) to give the approxi-
probable, which is consistent with the proposed biosynthesis
mate arrangement for the bound substrates of ScbA. The con-
model of butyrolactones (Sakuda et al., 1992).
served parts of the N- and C-terminal domains (the central a-helices and the preceding loops) are depicted in blue and dark
Using the 3D structures determined for FabA and FabZ, a
red, respectively. The side-chains of the conserved E78 and
structure of ScbA is proposed (Fig. 4). FabA and FabZ are
E240 are in cyan, the conserved Q82 and Q244 are in
homodimers (Leesong et al., 1996), and their two identical
magenta, and the conserved R81 and R243 are in red. Further
active sites are located at the dimer interface. We can assume
residues with potential functional importance are R228 (yellow)
that the two domains of ScbA will arrange in a similar
and D72 (orange).
fashion to form functional active sites. As the two domainshave diverged considerably (pairwise sequence identity of20 %), whereas the key residues remain unchanged (Fig. 1),
genome sequence of S. avermitilis, a similar multi-domain
it is probable that they catalyse similar reactions, but use
gene (SAV7361), which also has a FabA-homologous
domain, was found, as well as an individual FabA(SAV3654, 31 % amino acid identity to E. coli FabA) and
As ScbA has similarity to the active sites of fatty acid
FabZ (SAV3655, 31 % amino acid identity) homologue.
synthases, could ScbA also affect synthesis of fatty acids?
However, there are no other homologues of FabA or FabZ in
Streptomyces produces unsaturated fatty acids (Gesheva
S. coelicolor. Instead, we have identified a FabM homologue
et al., 1997), but, so far, from the S. coelicolor genome
(SCO5144), which is also highly conserved in S. avermitilis
sequence, no obvious type II FabZ homologue has been
(SAV3120, 86 % amino acid identity to SCO5144). In
identified (Bentley et al., 2002), while a distant orthologue
Streptococcus pneumoniae, a FabA homologue has not been
has been found in Streptomyces avermitilis (Ikeda et al.,
identified, but a FabM homologue that encodes a trans-2,
2003). Preliminary data suggest that S. coelicolor wild-type
cis-3 decenoyl-ACP isomerase has been found, along with a
and the scbA mutant produce C14, C16 and C17 unsaturated
FabZ homologue (Marrakchi et al., 2002). It is probable that
fatty acid synthesis in SMMS (data not shown). This result
SCO5144 is involved in the isomerase activity, but it is not
may exclude the possibility of ScbA involvement in
clear what gene in S. coelicolor is responsible for the
unsaturated fatty acids. However, the enzymes responsible
dehydration of b-hydroxyacyl-ACP to produce branched
for the synthesis of these fatty acids, i.e. the FabA or FabZ
fatty acids.
equivalent in S. coelicolor, have not been identified. Adomain within a large multi-domain gene (SCO0127) has
scbA is divergent to the c-butyrolactone receptor gene
been found to have a weak similarity to FabZ. From the
scbR. SCO6264, located adjacent to scbR, also affects
ScbA active sites are similar to FabA/Z
c-butyrolactone synthesis in S. coelicolor (T. Nihira & E.
function and the enzymic functions of ScbA, and this
Takano, unpublished). This gene is homologous to barS1,
strengthens the possibility that ScbA has dual function.
which is responsible for the reduction of the C-6 positionof VBs in S. virginiae (Shikura et al., 2002). barS1 is alsopositioned close to barX on the chromosome; barX is
NOTE ADDED IN PROOF
the scbA homologue in S. virginiae. There were 10 scbA
After this manuscript was accepted, a paper describing the in
homologues, including scbA and barX, found in the NCBI
vitro synthesis of A-factor, a c-butyrolactone produced by S.
database. Of these genes, nine have surrounding sequences
griseus, was published (Kato et al., 2007).
available for analysis, and six of these possess homologousgenes that probably encode members of the short-chaindehydrogenase family of proteins, which includes SCO6264
and the barS1 gene product. The genes adjacent to the afsA/
We thank K. Chater and A. Hesketh for critical reading of the
scbA-like gene mmfL in S. coelicolor plasmid SCP1 and avaA
manuscript, T. Ha¨rtner for the fatty acid analysis, and T. Nihira for
in S. avermitilis, mmfH and SAV2267, respectively, are
personal communication. N.-H. H. was funded by the Deutsche
classified as medium-chain acyl-CoA dehydrogenases, and
Forschungsgemeinschaft (TA428/1-1, 1-2).
are somewhat distant from the others, but still havehomology to dehydrogenases. The only exception to the‘rule' of having a dehydrogenase homologue in the cluster
was S. griseus. This species is atypical, as the c-butyrolactone
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z.,
receptor is at least 100 kb away from afsA, which is the scbA
Miller, W. & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new
homologue (Ando et al., 1997). Considering the conserva-
generation of protein database search programs. Nucleic Acids Res 25,
tion of the position of these dehydrogenase homologues, it is
possible that these genes are also involved in c-butyrolactone
Ando, N., Matsumori, N., Sakuda, S., Beppu, T. & Horinouchi, S.
synthesis, as in the case of barS1 and SCO6264. The
(1997). Involvement of AfsA in A-factor biosynthesis as a key
differences in the dehydrogenase family may also be related
enzyme. J Antibiot 50, 847–852.
to differences in the c-butyrolactone precursor and end-
Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A. M., Challis, G. L.,
product structures.
Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H. &other authors (2002). Complete genome sequence of the model
We have previously shown that the scbA deletion mutant
actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147.
overproduces antibiotics (Takano et al., 2001). In the
Bierman, M., Logan, R., O'Brien, K., Seno, E. T., Rao, R. N. &
present experiment, we were able to detect a slight difference
Schoner, B. E. (1992). Plasmid cloning vectors for the conjugaltransfer of DNA from Escherichia coli to Streptomyces spp. Gene 116,
in antibiotic production for all the strains using the
integrative plasmid. This may be due to the integration of
Brock, D. J., Kass, L. R. & Bloch, K. (1967). b-hydroxydecanoyl
pSET152, which affects the production of the pigmented
thioester dehydrase. II. Mode of action. J Biol Chem 242, 4432–4440.
antibiotics, both positively and negatively; we observed this
Camilli, A. & Bassler, B. L. (2006). Bacterial small-molecule signaling
repeatedly even in the wild-type with the vector alone, and
pathways. Science 311, 1113–1116.
also the instability of the vector integration without
Chiu, J., March, P. E., Lee, R. & Tillett, D. (2004). Site-directed, ligase-
selection (data not shown). To overcome this problem,
RT-PCR was conducted on both the regulator and the
approaching 100 % efficiency in 4 h. Nucleic Acids Res 32, e174.
biosynthesis genes. However, no difference was observed in
Chung, C. T., Niemela, S. L. & Miller, R. H. (1989). One-step
the expression of the biosynthesis genes in any of the strains,
preparation of competent Escherichia coli: transformation and
and the expression of the regulators was variable. This is
storage of bacterial cells in the same solution. Proc Natl Acad Sci
possibly due to the RNA isolated at a relatively late time
U S A 86, 2172–2175.
point, which was too late to see a clear difference in the
Flett, F., Mersinias, V. & Smith, C. P. (1997). High efficiency
expression levels of the biosynthesis genes, as well as the
intergeneric conjugal transfer of plasmid DNA from Escherichia coli
effect of the integrated vector. To clearly determine the effect
to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155,223–229.
of the mutation on antibiotic production, we are currently
Rachev, R. & Bojkova, S. (1997).
introducing these mutations in ScbA into the chromosome.
composition of Streptomyces hygroscopicus strains producing anti-biotics. Lett Appl Microbiol 24, 109–112.
We have also previously reported a possible regulatoryfunction for ScbA (Takano et al., 2001), based on the
Hara, O. & Beppu, T. (1982). Mutants blocked in streptomycinproduction in Streptomyces griseus – the role of A-factor. J Antibiot
observation that expression of scbA was not detected in the
35, 349–358.
scbA mutant, and was not complemented by addition of
Heath, R. J. & Rock, C. O. (1996). Roles of the FabA and FabZ b-
SCB1. In the present work, the mutated ScbA proteins did
hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty
not produce bioactive SCB proteins, but their expression
acid biosynthesis. J Biol Chem 271, 27795–27801.
under their own promoter, albeit in the WC31 attachment
Horinouchi, S., Nishiyama, M., Suzuki, H., Kumada, Y. & Beppu, T.
site, was restored to the same level as the wild-type. This
The cloned Streptomyces bikiniensis (griseus) A-factor
suggests that we have now uncoupled the regulatory
determinant. J Antibiot 36, 636–641.
N.-H. Hsiao and others
Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H.,
Sakuda, S., Higashi, A., Nihira, T. & Yamada, Y. (1990). Biosynthesis
Shiba, T., Sakaki, Y., Hattori, M. & Omura, S. (2003). Complete
of virginiae butanolide A. J Am Chem Soc 112, 898–899.
genome sequence and comparative analysis of the industrial
Sakuda, S., Higashi, A., Tanaka, S., Nihira, T. & Yamada, Y. (1992).
microorganism Streptomyces avermitilis. Nat Biotechnol 21, 526–531.
Biosynthesis of virginiae butanolide A, a butyrolactone autoregulator
Kato, J. Y., Funa, N., Watanabe, H., Ohnishi, Y. & Horinouchi, S.
from Streptomyces. J Am Chem Soc 114, 663–668.
(2007). Biosynthesis of c-butyrolactone autoregulators that switch on
Sakuda, S., Tanaka, S., Mizuno, K., Sukcharoen, O., Nihira, T. &
Yamada, Y. (1993). Biosynthetic studies on virginiae butanolide A, a
Streptomyces. Proc Natl Acad Sci U S A 104, 2378–2383.
butyrolactone autoregulator from Streptomyces. Part 2. Preparation
Kawachi, R., Akashi, T., Kamitani, Y., Sy, A., Wangchaisoonthorn, U.,
of possible biosynthetic intermediates and conversion experiments in
Nihira, T. & Yamada, Y. (2000). Identification of an a AfsA
a cell-free system. J Chem Soc Perkin Trans 1, 2309–2315.
homologue (BarX) from Streptomyces virginiae as a pleiotropic
Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning:
a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold
biosynthesis and virginiamycin M1 resistance. Mol Microbiol 36,
Spring Harbor Laboratory.
Shikura, N., Yamamura, J. & Nihira, T. (2002). barS1, a gene for
Khokhlov, A. S., Tovarova, I. I., Borisova, L. N., Pliner, S. A.,
biosynthesis of a c-butyrolactone autoregulator, a microbial
Shevchenko, L. A., Schevchenko, L. N., Kornitskaia, EIa., Ivkina,
signalling molecule eliciting antibiotic production in Streptomyces
N. S. & Rapoport, I. A. (1967). The A-factor, responsible for
species. J Bacteriol 184, 5151–5157.
streptomycini. Dokl Akad Nauk SSSR 177, 232–235.
So¨ding, J., Biegert, A. & Lupas, A. N. (2005). The HHpred interactiveserver for protein homology detection and structure prediction.
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A.
Nucleic Acids Res 33, W244–248.
Practical Streptomyces Genetics. Norwich: John Innes
Sonnhammer, E. L., Eddy, S. R., Birney, E., Bateman, A. & Durbin, R.
(1998). Pfam: multiple sequence alignments and HMM-profiles of
Kimber, M. S., Martin, F., Lu, Y., Houston, S., Vedadi, M., Dharamsi, A.,
protein domains. Nucleic Acids Res 26, 320–322.
Fiebig, K. M., Schmid, M. & Rock, C. O. (2004). The structure of(3R)-hydroxyacyl-acylcarrier
Strauch, E., Takano, E., Baylis, H. A. & Bibb, M. J. (1991). The
Pseudomonas aeruginosa. J Biol Chem 2793, 52593–52602.
stringent response in Streptomyces coelicolor A3(2). Mol Microbiol 5,289–298.
Kostrewa, D., Winkler, F. K., Folkers, G., Scapozza, L. & Perozzo, R.
(2005). The crystal structure of PfFabZ, the unique b-hydroxyacyl-
Takano, E. (2006). c-Butyrolactones: Streptomyces signaling mole-
ACP dehydratase involved in fatty acid biosynthesis of Plasmodium
cules regulating antibiotic production and differentiation. Curr Opin
falciparum. Protein Sci 14, 1570–1580.
Microbiol 9, 287–294.
Leblond, P., Fischer, G., Francou, F. X., Berger, F., Gue´rineau, M. &
Takano, E., Chakaraburtty, R., Nihira, T., Yamada, Y. & Bibb, M. J.
Decaris, B. (1996). The unstable region of Streptomyces ambofaciens
(2001). A complex role for the c-butyrolactone SCB1 in regulating
includes 210 kb terminal inverted repeats flanking the extremities of
antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol
the linear chromosomal DNA. Mol Microbiol 19, 261–271.
41, 1015–1028.
Leesong, M., Henderson, B. S., Gillig, J. R., Schwab, J. M. & Smith,
Takano, E., Kinoshita, H., Mersinias, V., Bucca, G., Hotchkiss, G.,
J. L. (1996). Structure of a dehydratase-isomerase from the bacterial
Nihira, T., Smith, C. P., Bibb, M., Wohlleben, W. & Chater, K. (2005).
pathway for biosynthesis of unsaturated fatty acids: two catalytic
A bacterial hormone (the SCB1) directly controls the expression of a
activities in one active site. Structure 4, 253–264.
pathway-specific regulatory gene in the cryptic type I polyketidebiosynthetic gene cluster of Streptomyces coelicolor. Mol Microbiol 56,
MacNeil, D. J., Occi, J. L., Gewain, K. M., MacNeil, T., Gibbons, P. H.,
Ruby, C. L. & Danid, S. J. (1992). Complex organization of theStreptomyces avermitilis genes encoding the avermectin polyketide
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M. & Hardie, K. R.
synthetase. Gene 155, 119–125.
(2005). Making ‘sense' of metabolism: autoinducer-2, LuxS andpathogenic bacteria. Nature Rev Microbiol 3, 383–396.
Marrakchi, H., Choi, K. H. & Rock, C. O. (2002). A new mechanismfor anaerobic unsaturated fatty acid formation in Streptococcus
Venturi, V. (2006). Regulation of quorum sensing in Pseudomonas.
pneumoniae. J Biol Chem 277, 44809–44816.
FEMS Microbiol Rev 30, 274–291.
Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995).
Yamada, Y. (1999). Autoregulatory factors and regulation of
SCOP: a structural
classification of proteins database for the
antibiotic production in Streptomyces. In Microbial Signalling and
investigation of sequences and structures. J Mol Biol 247, 536–540.
Communication (Society for General Microbiology Symposium no.
57), pp. 177–196. Edited by R. R. England, G. Hobbs, N. J. Bainton
Paget, M. S. B., Chamberlin, L., Atrih, A., Foster, S. J. & Buttner, M. J.
& D. McL. Roberts. Cambridge: Cambridge University Press.
(1999). Evidence that the extracytoplasmic function sigma factor sEis required for normal cell wall structure in Streptomyces coelicolorA3(2). J Bacteriol 181, 204–211.
Edited by: J. Anne´
Source: http://www.soeding.genzentrum.lmu.de/assets/Lab-Soeding/Lab-Soeding-Publications/Microbiol-2007-Hsiao-Takano-ScbA.pdf
Plan Performance -Executive Summary - Municipal P ension Plan of British Columbia Plan Performance Review™ Executive Summary Table PLAN PERFORMANCE PERFORMANCE Latest 24 Month Trending FV SAVINGS 36 MONTHS ¹ General Plan Metrics Total Plan Spending Total Amount Paid ($) by Plan
Papier aus chlorfrei gebleichtem Zellstoff Jahrg. 28 . Nr. 225 Informationsdienst für Neurologen und Psychiater Morbus ParkinsonWird M. Parkinson durch eine Neuromelanin-stimulierte Immun- „Vor 50 Jahren wurden koreanische Ein Abbild unserer Gesellschaft antwort hervorgerufen? Krankenschwestern angeworben: Jetzt kommen die Chinesen" Eine zukunftsträchtige Disziplin









